Hello,
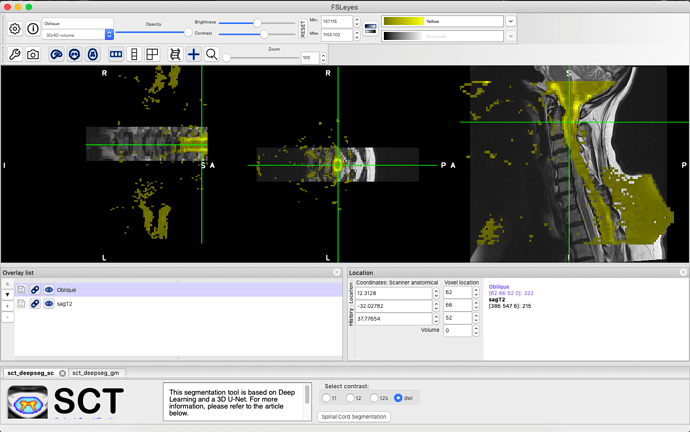
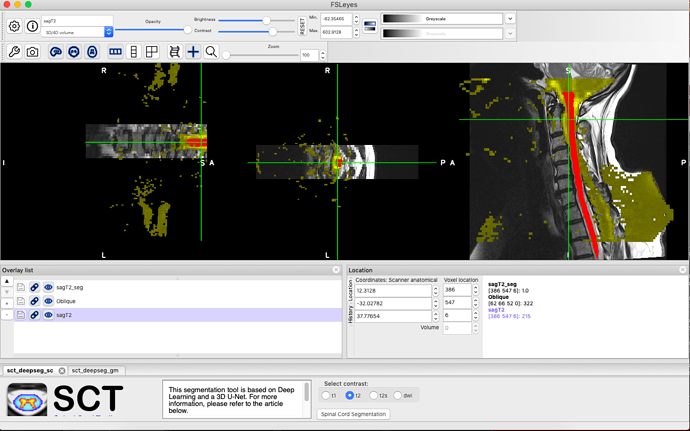
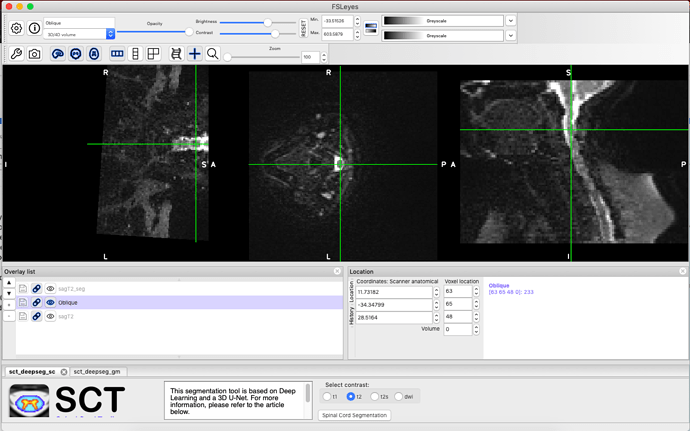
I am trying to find a program to perform spine image processing for the spinal DTI scans. Specifically, I need to perform a non-rigid registration to straighten the spinal cord in the DWI Oblique sequence to align with the other sequences in the MRI scan (T1, T2). I attempted to follow the workshop pdfs but cannot see how to do this when I cannot visualise the vertebral column for labeling in the DTI sequence for eventual warping. I have experience with MRtrix and 3D slicer, but had no luck performing registration in 3D slicer. Below are a couple screenshots showing the DWI sequence (yellow) and the T2 sequence w/ segmentation (red), and the DWI sequence in grey scale. Any guidance would be helpful. Thank you.
Hi @Caleb_Stewart,
The vertebral labeling comes from the anatomical data, and the registered PAM50 template is then warped to the DWI scan via the anatomical data. Here is an example of commands (using SCT example dataset):
# Download example data
sct_download_data -d sct_example_data
cd sct_example_data/t2
# Segment spinal cord
sct_deepseg_sc -i t2.nii.gz -c t2 -qc ~/qc-dwi
# Label vertebrae
sct_label_vertebrae -i t2.nii.gz -s t2_seg.nii.gz -c t2 -qc ~/qc-dwi
# Generate labels at C3 and T2
sct_label_utils -i t2_seg_labeled.nii.gz -vert-body 3,9 -o labels_vert.nii.gz
# Register to PAM50
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -l labels_vert.nii.gz -c t2 -qc ~/qc-dwi
# Go to dmri folder, create mask and perform motion correction
cd ../dmri/
sct_maths -i dmri.nii.gz -mean t -o dmri_mean.nii.gz
sct_register_multimodal -i ../t2/t2_seg.nii.gz -d dmri_mean.nii.gz -identity 1 -x nn
sct_create_mask -i dmri_mean.nii.gz -p centerline,t2_seg_reg.nii.gz -size 35mm
sct_crop_image -i dmri.nii.gz -m mask_dmri_mean.nii.gz -o dmri_crop.nii.gz
sct_dmri_moco -i dmri_crop.nii.gz -bvec bvecs.txt
# Segment cord on DWI data
sct_deepseg_sc -i dmri_crop_moco_dwi_mean.nii.gz -c dwi -qc ~/qc-dwi
# Register DWI with anatomical data (use segmentations for more robustness)
sct_register_multimodal -i dmri_crop_moco_b0_mean.nii.gz -iseg dmri_crop_moco_dwi_mean_seg.nii.gz -d ../t2/t2.nii.gz -dseg ../t2/t2_seg.nii.gz -param step=1,type=seg,algo=centermass:step=2,type=seg,algo=bsplinesyn,metric=MeanSquares,smooth=1,slicewise=1,iter=3 -qc ~/qc-dwi
Below is the result:
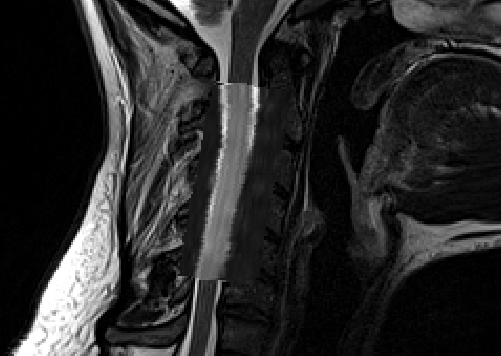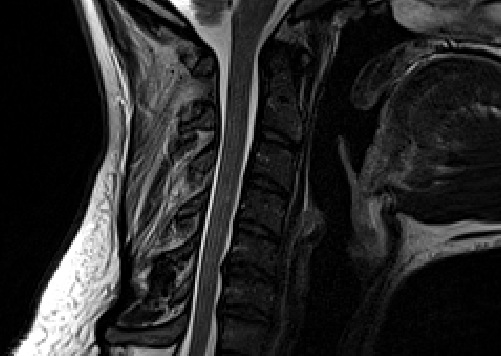
And here is the QC report: qc-dwi.zip (2.4 MB)
Note that we usually recommend to register the DWI scans with the PAM50 (not the anatomical data), and then warp the PAM50 atlases onto the DWI space and then perform analyses in the DWI space. This minimizes interpolation errors when bringing your DWI scans (or results) into the anatomical space. You can find the steps in the example batch_processing.sh.
Cheers,
Julien
1 Like
Hi @Caleb_Stewart,
Based on our email conversation where you sent me your data, below is a list of commands that are more tailored to your data and what you would like to achieve:
# Resample to 0.6 mm iso for robust cord segmentation
sct_resample -i t2_sag.nii.gz -mm 0.6x0.6x0.6 -o t2_sag_r.nii.gz
# Segment spinal cord on T2 data. Note: this segmentation is very approximate because the R-L resolution is poor (sagittal image)
sct_deepseg_sc -i t2_sag_r.nii.gz -c t2 -qc qc
# Separate DWI from b=0 images and average them
sct_dmri_separate_b0_and_dwi -i dmri.nii.gz -bvec dmri.bvec -a 1
# Segment spinal cord on the b=0 data
sct_deepseg_sc -i dmri_b0_mean.nii.gz -c t2 -qc qc
# Dilate the segmentation because due to the strong artifacts, some slices have no segmentation, which will make the slicereg algorithm perform poorly
sct_maths -i dmri_b0_mean_seg.nii.gz -dilate 3 -o dmri_b0_mean_seg_dil3.nii.gz
# Register dmri to T2. Note: Due to the very large distortions, the trick here is to allow for more deformation by setting smooth=5 (default: 0).
sct_register_multimodal -i dmri_b0_mean.nii.gz -iseg dmri_b0_mean_seg_dil3.nii.gz -d t2_sag_r.nii.gz -dseg t2_sag_r_seg.nii.gz -param step=1,type=seg,algo=slicereg,smooth=5:step=2,type=im,algo=syn,metric=CC,smooth=0,slicewise=0,iter=3 -qc qc
Below are the registration results. Left: before registration, right: after registration.
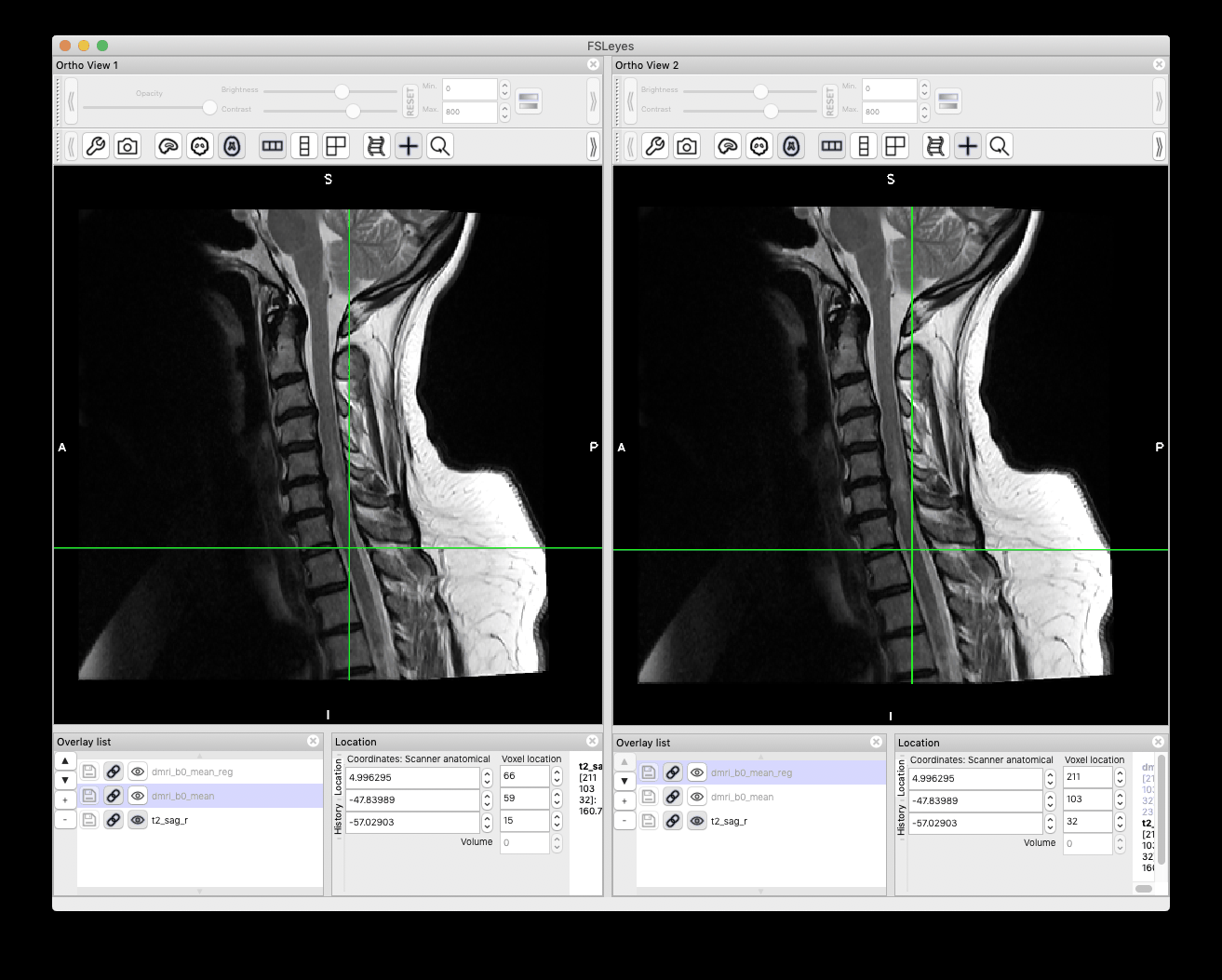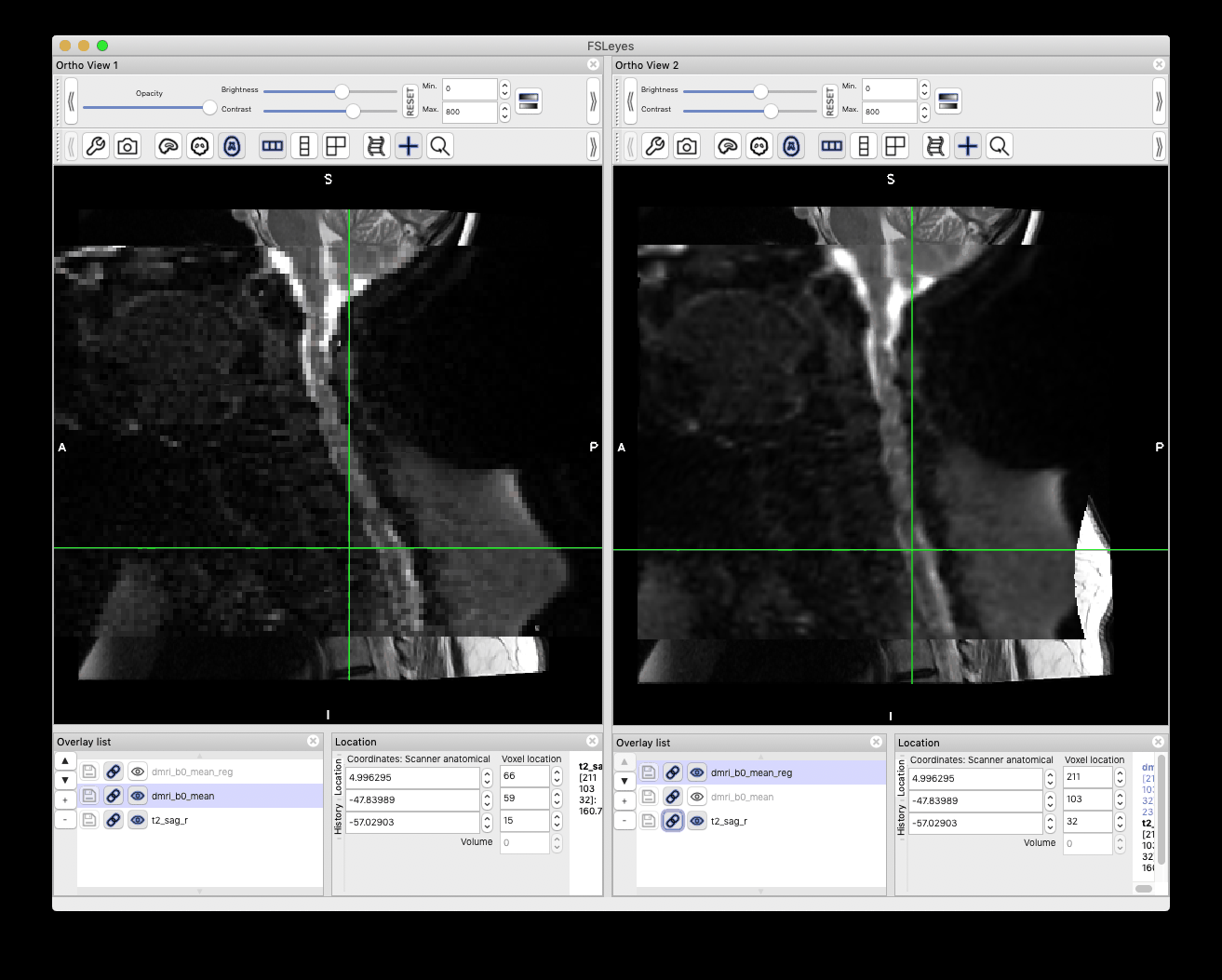
Best,
Julien
1 Like