Dear Dr Cohen-Adad,
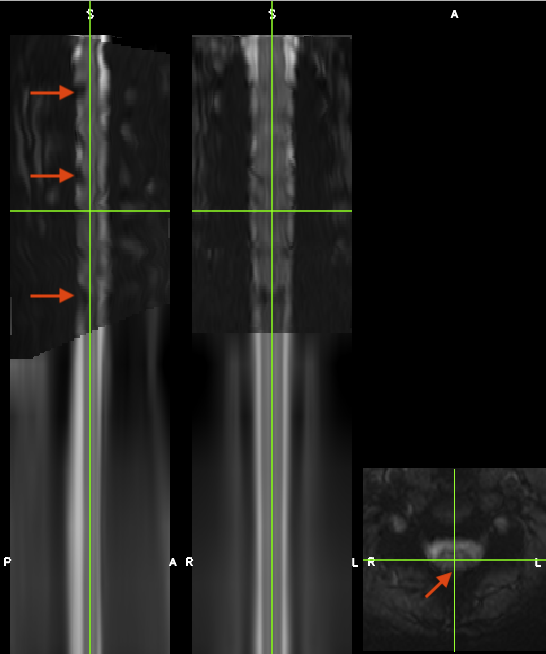
I am trying to register my EPIs to PAM50_t2 template (indeed registering PAM50 to my EPI).
For that, I have used a different number of parameters, with different -algo parameters etc. For initwarp I used the warp of my participant T2 to PAM50 template (which seemed to work very well). The segmentations of T2 and EPI are pretty good as well, I manually drew the segmentation of EPI.
However, unfortunately, I am still not able to find the optimal set of parameters.
In addition to that, I also have tried a 2 step strategy where I registered individual T2 and EPI and T2 to PAM50 template, concatenated the transformations and applied the final/combined warp to the EPI.
In the end, this seems to work a bit better compared to the sct_register_multimodal with different parameters I have tried but still, I am still not sure about the results.
Here are some parameters I have tried for register multimodal (and also 2 step approach which I do not copy here):
sct_register_multimodal -i /home/raid2/mkaptan/sct_4.0.2/data/PAM50/template/PAM50_t2.nii.gz -d EPI.nii.gz -iseg /home/raid2/mkaptan/sct_4.0.2/data/PAM50/template/PAM50_cord.nii.gz -dseg EPI_seg.nii.gz -param step=1,type=seg,algo=centermass:step=2,type=seg,algo=bsplinesyn,metric=MeanSquares,smooth=1,slicewise=1,iter=3 -initwarp warp_template2anat.nii.gz -initwarpinv warp_anat2template.nii.gz -x spline
sct_register_multimodal -i /home/raid2/mkaptan/sct_4.0.2/data/PAM50/template/PAM50_t2.nii.gz -d EPI.nii.gz -iseg /home/raid2/mkaptan/sct_4.0.2/data/PAM50/template/PAM50_cord.nii.gz -dseg EPI_seg.nii.gz -param step=1,type=seg,algo=slicereg,smooth=3:step=2,type=seg,algo=bsplinesyn,slicewise=1,iter=3 -initwarp warp_template2anat.nii.gz -initwarpinv warp_anat2template.nii.gz -x spline
sct_register_multimodal -i /home/raid2/mkaptan/sct_4.0.2/data/PAM50/template/PAM50_t2.nii.gz -d EPI.nii.gz -iseg /home/raid2/mkaptan/sct_4.0.2/data/PAM50/template/PAM50_cord.nii.gz -dseg EPI_seg.nii.gz -param step=1,type=seg,algo=slicereg,smooth=3 -initwarp warp_template2anat.nii.gz -initwarpinv warp_anat2template.nii.gz -x spline
sct_register_multimodal -i /home/raid2/mkaptan/sct_4.0.2/data/PAM50/template/PAM50_t2.nii.gz -d EPI.nii.gz -iseg home/raid2/mkaptan/sct_4.0.2/data/PAM50/template/PAM50_cord.nii.gz -dseg EPI_seg.nii.gz -param step=1,type=seg,algo=centermass,smooth=3 -initwarp warp_template2anat.nii.gz -initwarpinv warp_anat2template.nii.gz -x spline
sct_register_multimodal -i /home/raid2/mkaptan/sct_4.0.2/data/PAM50/template/PAM50_t2.nii.gz -d EPI_cropped.nii.gz -iseg /home/raid2/mkaptan/sct_4.0.2/data/PAM50/template/PAM50_cord.nii.gz -dseg EPI_seg.nii.gz -param step=1,type=seg,algo=centermass:step=2,type=seg,algo=bsplinesyn,metric=MeanSquares,iter=10 gradStep=1:step=3,type=im,algo=syn,metric=CC,iter=20 -initwarp warp_template2anat.nii.gz -initwarpinv warp_anat2template.nii.gz -x spline
Could you please help me with that?
I am not able to upload the images due to size here but I can send via email if you would like to!
Thank you very much,
Best,
Merve