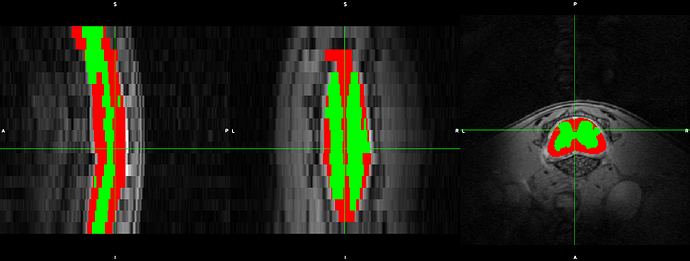
Hi,
Thanks for your help.
we are trying to do segmentation for mouse lumbar spinal cord following the instruction in the website below:
-
we did not do (sct_crop) cropping because we use cryocoil – signal is already quite limited to the spinal cord only
-
sct_propseg -i T1_440.nii.gz -c t2 -init-centerline viewer -rescale 3
We used -c t2 because the image contrast is more like t2 (we used T1/T2* gradient echo sequence)
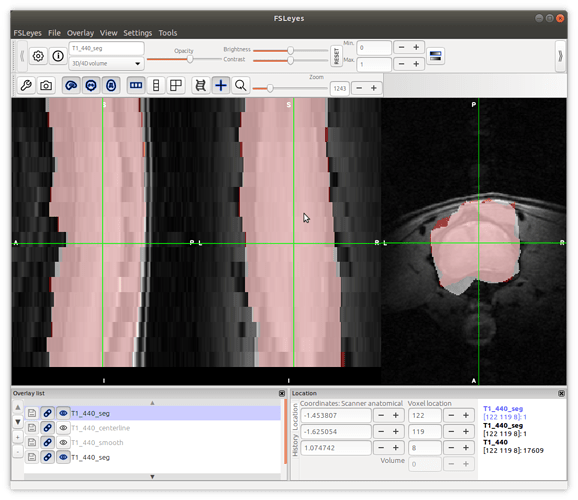
We noted that some parts of the cords were not included, but at some slices the muscles were covered.
Secondly, we noted that the _centerline does not stay in the centre of the cord. It deviates further away caudal of the slices.
-
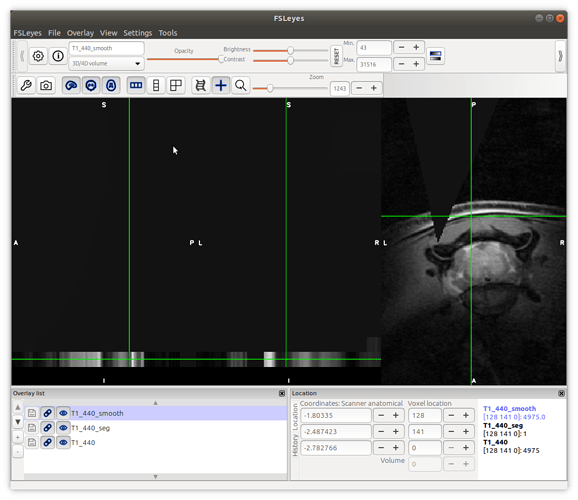
sct_smooth_spinalcord -i T1_440.nii.gz -s T1_440_centerline.nii.gz -smooth 5
this did not give very good results… all of the slices except the last slice have disappeared from the image?
-
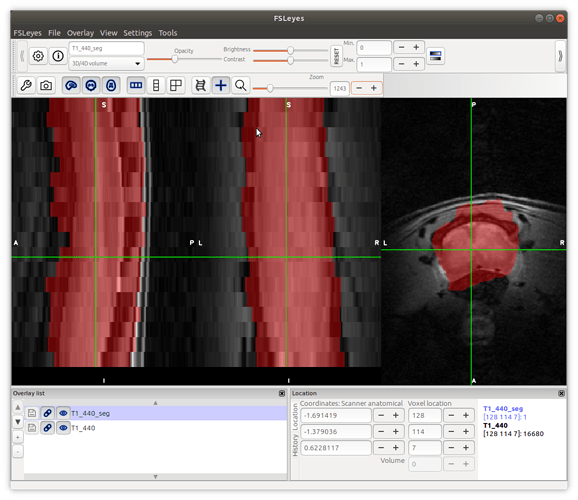
sct_propseg -i T1_440.nii.gz -c t2 -init-centerline T1_440_centerline.nii.gz -rescale 3
we did not use the smoothed image result for this step because the result from step 3 was not good.
Therefore we used the original image.
I think the results are similar to that in step 2: the segmentation is sometimes larger or smaller than the cord at different slices…
please have a look on the screenshots.
Best regards,
Abdullah
Hi Abdullah,
The SCT models were trained with data that look quite different than the contrast you used here. Do you have other contrasts (e.g., T2, DWI, etc.) that we can try the segmentation on? If so, would you be able to share them with me so I can try on my end?
If not, follow-up questions are:
- how many of these volumes do you need to analyze?
- are you interested only in spinal cord segmentation or also GM segmentation?
Depending on your answers, we could look into training a model specific for your data. That should only take 1-2 hours and will save you a lot of time (depending on how many datasets you need to analyze).
Cheers,
Julien
Thanks Julien,
Yes we have DWI as a raw image and as processed FA, AD, MD and RD.
And we love to share them with you.
For segmentation we want WM and GM and whole cord for over 40 mice that we currently have.
Best regards,
Abdullah
Ok, 40 mice is definitely worth training a new model. I guess segmentation could be done on the T1w and then you would register the DWI data on the T1w to use the T1w seg for the DWI, right?
If you can share let’s say T1w dataset from 2 mice, along with WM and GM segmentations (only on 3 slices per mice), we will train a new model and you can the apply it on the other mice.
Exactly this what we want.
But is it OK to do the segmentation for three slices but they have one or two slice gap between them i.e slice #3, 5, 7?
because the shape of WM/GM changes from caudal to rostral.
Best regards,
Abdullah
The idea is to pick a subset of slices that are most representative of all your slices
Julien,
I sent the files through the email yesterday but now I realised it did not reach you.
sorry for this.
T1_441.nii.gz (1.8 MB) T1_Seg_SCT_441.nii.gz (3.7 KB) T1_440.nii.gz (1.8 MB) T1_Seg_SCT_440.nii.gz (3.5 KB)
Perfect! let us try and come back to you.
Hi Charley,
Sorry for the delay I did not see your email!
I attached five data set one of them diseased (EAE_3) hopefully it helped much better.
I really appreiate your help.
Cheers
Abdullah Althobity
T1_437.nii.gz (1.9 MB) T1_438.nii.gz (1.8 MB) T1_439.nii.gz (1.8 MB) T1_442.nii.gz (1.9 MB) T1_EAE3.nii.gz (1.7 MB) T1_seg_SCT_437.nii.gz (3.4 KB) T1_seg_SCT_438.nii.gz (3.7 KB) T1_seg_SCT_439.nii.gz (3.3 KB) T1_seg_SCT_442.nii.gz (3.5 KB) T1_seg_SCT_EAE_3.nii.gz (4.5 KB)
1 Like
Hi Abdullah,
Many thanks for your efforts and sorry for the late reply.
We will be back to you soon.
Cheers,
Charley
Hi Charley,
I hope you and Julien are doing well in this crisis!
I Just would like to remind you if have done the training model?
Best regards,
Abdullah
Hi Abdullah,
Yes, the model is trained. We are currently implementing it via a new SCT command (sct_deepseg), see PR #2639, which you will be the first to test! I expect it to be ready within 1 week-- stay tuned!
1 Like
Hi @Abdullah_Althubaiti,
We are very sorry for the delay, but it was a lot of work to put everything together. We are happy to let you know that a development version is now ready to use. Below are the instructions:
# If you are using a release version, you will need to install another SCT version from git. Otherwise, skip that line below
git clone https://github.com/neuropoly/spinalcordtoolbox.git sct-dev
cd sct-dev
# Update git repos
git fetch origin
# Checkout prototype branch
git checkout -b jca/2626-deepseg origin/jca/2626-deepseg
# Install SCT. Note: after the installation you will get a message that says "Installation failed". You can ignore this message. The installation actually succeeded (we are still working on this message)
yes | ./install_sct
# VERY IMPORTANT: After the installation, open a new Terminal (to load updated environment variables)
# Then, install mice models
sct_deepseg -install-model uqueensland_mice_sc
sct_deepseg -install-model uqueensland_mice_gm
# Go to the folder where you have your MRI data
# Segment spinal cord and gray matter (replace with your image file name)
sct_deepseg -i T1_EAE3.nii.gz -m uqueensland_mice_sc
sct_deepseg -i T1_EAE3.nii.gz -m uqueensland_mice_gm -o _gmseg
Below is an example of segmentation results:
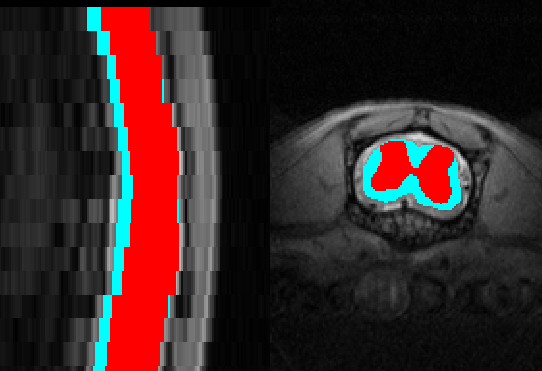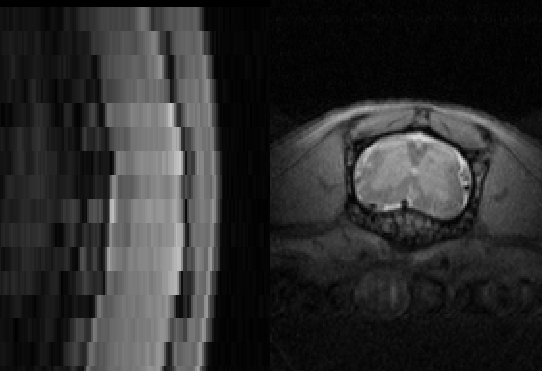
Please send us as much feedback as possible so we can improve this function before we release it.
Thank you @Charley_Gros and @anon33887296 for your help with this new feature!
Cheers,
Julien
Hi Julien,
Thank you very much.
much appreciate all your efforts and Charley and Nick.
Will work on it and will give you the updates.
Cheers,
Abdullah
2 Likes
Hi Julien & Charley,
Thanks for training the model and doing all this work.
I have installed SCT and everything thing went well, thanks to your clear guidance.
Please see the screenshot for current results
it looks fairly good but for grey matter it is a little tricky.
it could be because it is diseased mouse. (I also attached the same mouse “347” with its segmentation)
On other mice, specially the healthy ones looks much better with some errors.
I have attached one more sample (healthy one) with it segmentation on different locations hopefully it helps.
T1_67.nii.gz (1.8 MB)
T1_seg_SCT_67.nii.gz (3.4 KB)
T1_347.nii.gz (1.8 MB)
T1_seg_SCT_347.nii.gz (4.3 KB)
the total of new slices is 11, please if you need more slices/data let me know.
Cheers,
Abdullah
Hi Abdullah,
Thanks for the feedback. I’m wondering if we shouldn’t add a diseased mouse in the training instead of a healthy one, so that the model sees more images that currently have failed segmentations? @Charley_Gros?
Hi Abdullah,
Many thanks for the feedback and the new slices manually segmented.
I am going to add them to the training pipeline.
As Julien said: it would be great if you could segment few slices (e.g. 3 slices) on a diseased mouse. Please let us know if that’s something you could do.
We will come back to you shortly with a new model that you will be able to use on your data.
Cheers
Thanks both of you,
here is one diseased mouse with three segmented slices.
If you need more segmentations please just let me know.
Cheers
Abdullah
T1_seg_SCT_423.nii.gz (3.1 KB) T1_423.nii.gz (1.9 MB)
Many thanks Abdullah! We will come back to you shortly
MESSAGE EDIT: 12:07 15/05/2020 Brisbane Time
Hi Abdullah,
We have just updated the models. I believe the results are much better now.
Pull changes on the Github PR:
cd sct-dev
git pull origin jca/2626-deepseg
To get the new models, please run the following lines:
sct_deepseg -install-model mice_uqueensland_sc
sct_deepseg -install-model mice_uqueensland_gm
Then run your segmentation the same way:
sct_deepseg -i T1_EAE3.nii.gz -m mice_uqueensland_sc -o _scseg
sct_deepseg -i T1_EAE3.nii.gz -m mice_uqueensland_gm -o _gmseg
Please let me know if you have any issues and please let me know how the new models went
Cheers