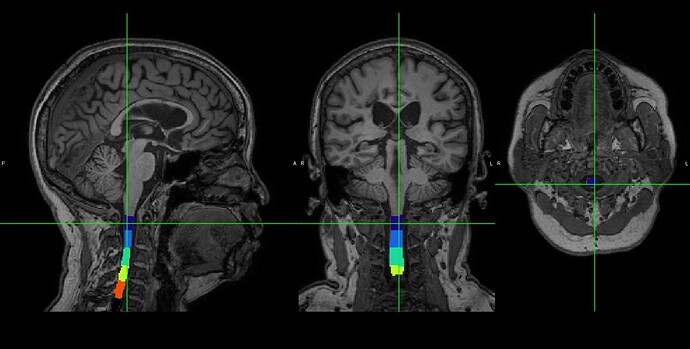
Dear SCT experts,
I have a T1 contrast brain image which included the upper cervical cord. I input the image to SCT. However, SCT can not identify the upper cervical cord from the image. May I know is the SCT only process spinal cord image?
You could download the log file and the T1 contrast brain image in the following link:
Hi,
If you wish to use propseg, you need to initialize the starting slice to be in the spinal cord with the flag -init. Example:
sct_propseg -i T1_contrast.nii -init 10 -c t1
Moreover, there is another segmentation tool based on deep learning, which gives slightly better segmentation results. Here is the command:
sct_deepseg_sc -i T1_contrast.nii -c t1
And here is the result:
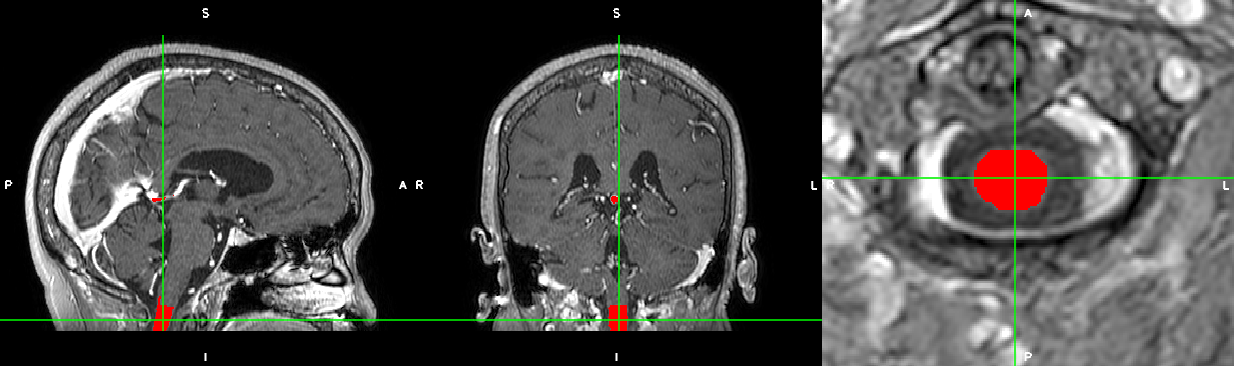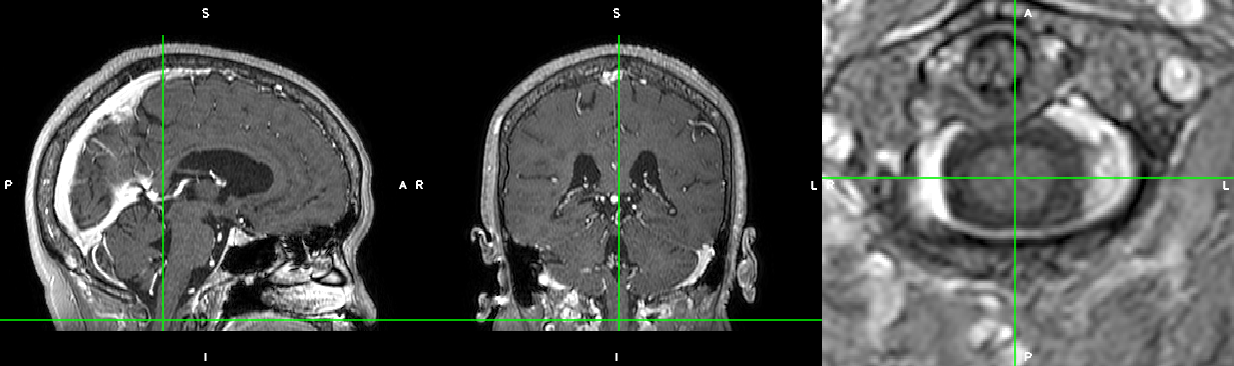
Cheers,
Julien
Thanks for your advises.
Is the “10” of " -init 10" is the coordinate of the image?
How could I obtain the corresponding number of starting slice of the spinal cord of my subject image?
Frank
Hi,
-init 10 tells the program to start the initialization at the 10th axial slice. So if all your subjects are more or less acquired with the same FOV, this should work fairly well.
Best,
Julien
Hi Julien,
Thank you for the further explain.
How could I obtain the volume of specific section of the tagged spinal cord after run the sct_propseg , such as the upper cervical cord?
Frank
Hi Frank,
Maybe this post will help?
Cheers
Julien
Hi Julien,
For the command of computing cord volume:
sct_analyze_lesion -m t2_seg_cervical.nii.gz -s t2_seg_cervical.nii.gz
How could I generate the “seg_cervical.nii.gz”?
Is it by using the “sct_label_utils” command?
Frank
Hi Frank,
To compute cord volume within a specific level, you would need to label the levels first, e.g. with sct_label_vertebrae. However, in your case, we don’t see any disc (FOV is to high up) so I don’t recommend this approach.
Moreover, it is important to realize that volume measurement is highly sensitive to the number of slices that are included in the calculation. E.g., if one subject shows 30 slices of the upper cervical cord and another subject shows 25 slices, then you will not be able to compare volumes properly, unless you normalize with the number of slices, which would be the equivalent of computing average CSA across slices-- which I recommend.
Best,
Julien
Dear Julien,
Thank you for your advise.
I speculated most of of my subjects, their image captured 4-5 discs of C1 & C2 cord.
Here is one of my subject:
I ran sct_label_vertebrae on this subject. How could I obtain a specific cord volume? My goal is obtaining the first 3cm of upper cervical cord.
Regards
Frank
Hi,
It is extremely important to understand that when computing cord volume defined between vertebral levels (e.g. C1-C3) this measure will be extremely dependent on the length of the vertebral bodies, which varies across individuals. If you were to compute the average CSA between C1-C3, you would not suffer from that bias.
So, before advising on how to do it, I want to make sure that the output value will not be misinterpreted.
Best,
Julien
Dear Julien,
So, this method is only suitable for measuring relatively long length of the spinal cord?
Because the difference in 1 disc is a relative proportion error when measuring the average CSA between C1-C3. Is my understanding right?
best,
Frank
So, this method is only suitable for measuring relatively long length of the spinal cord?
If your goal is to measure things like cord atrophy, volume should never be used. As you can see in the literature, in applications such as MS, what is measured is CSA (at a single level, or averaged across levels), not cord volume.
Dear Julien,
Sorry, for my misuse of wording.
I want obtain the average CSA of first 30mm of cervical cord. (The precise definition: selecting a fixed set of contiguous axial slices starting from the most superior point of C1 and ending at a position 30mm more inferior, where the 30mm length was measured perpendicular to the axial plane.)
As you said, it should suitable to use the SCT to obtain the average CSA after normalizing the number of slices included in the calculation.
If that is suitable, how could I perform this calculation of average CSA on my subject image?
Thanks for the detail explanation again.
Frank
hi,
it is currently not possible to aggregate metrics by specifying a distance along the cord centerline. I’ve opened an issue here, so we will make sure to add this functionality in the future.
However, for now, you could do two things: Compute average CSA:
- Across specified vertebral levels, or
- Across a specified number of slices. If you know your slice thickness, you can figure out how many slices you need to equivalent 30mm. However, this length will be impacted by the angle between the cord centerline and the vector orthogonal to the axial slice plane.
Below is an example for option 1:
# Segment spinal cord.
# Note: replace "t2" by your image file name and adapt flag -c depending on the image contrast.
sct_deepseg_sc -i t2.nii.gz -c t2 -qc qc
# Vertebral labeling
sct_label_vertebrae -i t2.nii.gz -s t2_seg.nii.gz -c t2 -qc qc
# Compute cross-sectional area and average between C1 and C3 levels
sct_process_segmentation -i t2_seg.nii.gz -vert 1:3 -o csa_C1-C3.csv