Hi @jcohenadad ,
I had a few unexpected results while I was trying to implement your code:
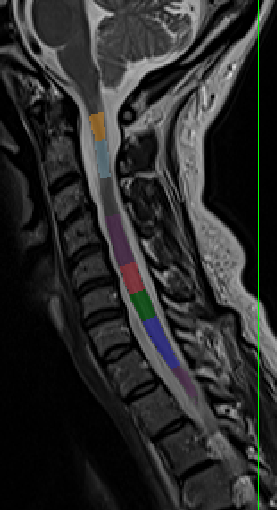
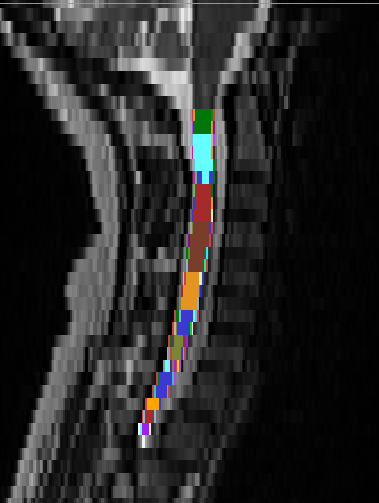
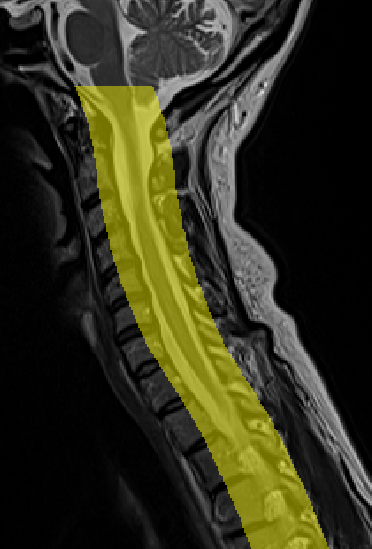
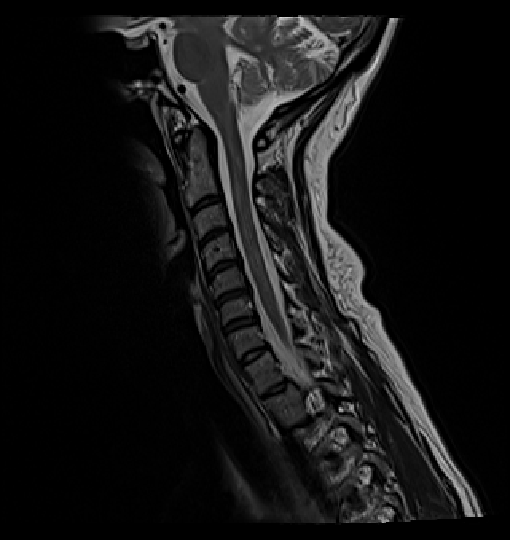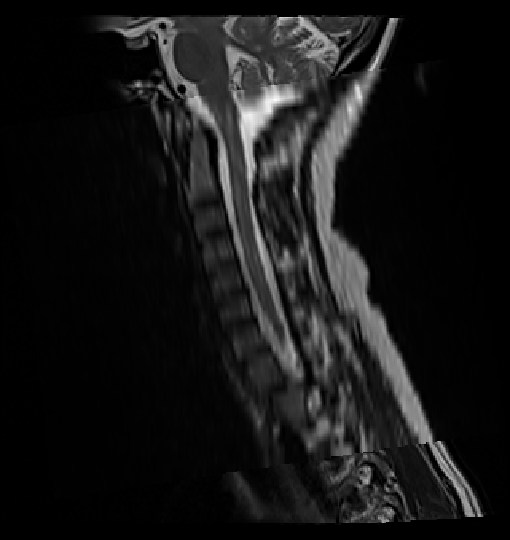
- When I run the following commands to get the vertebrae labels it has a problem detecting disk 5:
sct_deepseg_sc -i sag_cervical.nii.gz -c t2 -qc "$QCF"
sct_label_vertebrae -i sag_cervical.nii.gz -s sag_cervical_seg.nii.gz -c t2 -qc "$QCF"
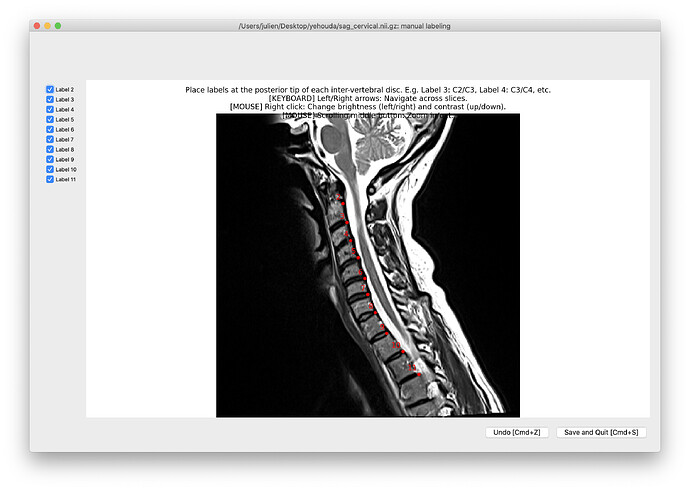
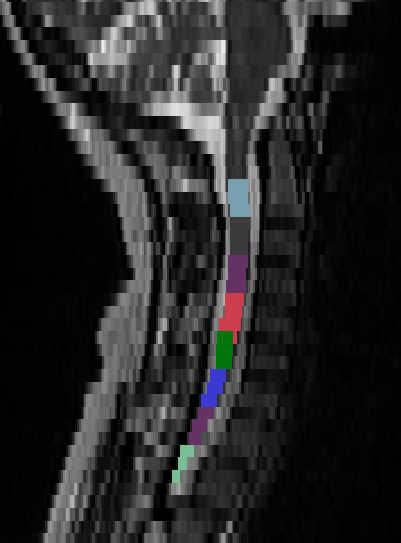
- When I tried to manually fix the labels with the following commands I got en error:
command:
sct_label_utils -i sag_cervical.nii.gz -create-viewer 3,4,5 -o labels_disc.nii.gz -qc "$QCF" -msg "Place labels at the posterior tip of each inter-vertebral disc. E.g. Label 3: C2/C3, Label 4: C3/C4, etc."
sct_label_vertebrae -i sag_cervical.nii.gz -s sag_cervical_seg.nii.gz -c t2 -qc "$QCF" -initlabel labels_disc.nii.gz
I got the error:
Get z and disc values from straight label...
.. [273, 0]
Look for template...
Path template: /home/yael/sct_5.4/data/PAM50
Open template and vertebral levels...
Disc values from template: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19]
Z-values for each disc: [963, 938, 907, 870, 833, 800, 769, 735, 692, 646, 600, 551, 500, 449, 396, 342, 289, 231, 168, 104, 79]
Distances between discs (in voxel): [25.0, 31.0, 37.0, 37.0, 33.0, 31.0, 34.0, 43.0, 46.0, 46.0, 49.0, 51.0, 51.0, 53.0, 54.0, 53.0, 58.0, 63.0, 64.0, 25.0]
Detect intervertebral discs...
Current disc: -1 (z=273). Direction: superior
.. Peak found: z=8 (correlation = 0.2358752778730007)
Traceback (most recent call last):
File "/home/yael/sct_5.4/spinalcordtoolbox/vertebrae/core.py", line 206, in vertebral_detection
approx_distance_to_next_disc = list_distance[list_disc_value_template.index(current_disc - 1)]
ValueError: -2 is not in list
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/home/yael/sct_5.4/spinalcordtoolbox/scripts/sct_label_vertebrae.py", line 485, in <module>
main(sys.argv[1:])
File "/home/yael/sct_5.4/spinalcordtoolbox/scripts/sct_label_vertebrae.py", line 422, in main
verbose=verbose, path_template=path_template, path_output=path_output, scale_dist=scale_dist)
File "/home/yael/sct_5.4/spinalcordtoolbox/vertebrae/core.py", line 208, in vertebral_detection
logger.warning('Disc value not included in template. Using previously-calculated distance: %s', approx_distance_to_next_disc)
UnboundLocalError: local variable 'approx_distance_to_next_disc' referenced before assignment
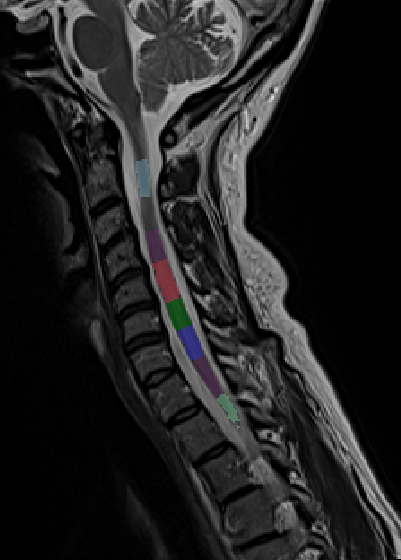
Same happened when I was trying to get the vertebrae labels for t2 sagittal stir scan:
sct_deepseg_sc -i sag_stir_cervical.nii.gz -c t2 -qc "$QCF"
sct_label_vertebrae -i sag_stir_cervical.nii.gz -s sag_stir_cervical_seg.nii.gz -c t2 -qc "$QCF" -initlabel labels_disc.nii.gz
I got the error:
Get z and disc values from straight label...
.. [295, 0]
Look for template...
Path template: /home/yael/sct_5.4/data/PAM50
Open template and vertebral levels...
Disc values from template: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19]
Z-values for each disc: [963, 938, 907, 870, 833, 800, 769, 735, 692, 646, 600, 551, 500, 449, 396, 342, 289, 231, 168, 104, 79]
Distances between discs (in voxel): [25.0, 31.0, 37.0, 37.0, 33.0, 31.0, 34.0, 43.0, 46.0, 46.0, 49.0, 51.0, 51.0, 53.0, 54.0, 53.0, 58.0, 63.0, 64.0, 25.0]
Detect intervertebral discs...
Current disc: -1 (z=295). Direction: superior
.. Peak found: z=-5 (correlation = 0.2045552821852658)
Traceback (most recent call last):
File "/home/yael/sct_5.4/spinalcordtoolbox/vertebrae/core.py", line 206, in vertebral_detection
approx_distance_to_next_disc = list_distance[list_disc_value_template.index(current_disc - 1)]
ValueError: -2 is not in list
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/home/yael/sct_5.4/spinalcordtoolbox/scripts/sct_label_vertebrae.py", line 485, in <module>
main(sys.argv[1:])
File "/home/yael/sct_5.4/spinalcordtoolbox/scripts/sct_label_vertebrae.py", line 422, in main
verbose=verbose, path_template=path_template, path_output=path_output, scale_dist=scale_dist)
File "/home/yael/sct_5.4/spinalcordtoolbox/vertebrae/core.py", line 208, in vertebral_detection
logger.warning('Disc value not included in template. Using previously-calculated distance: %s', approx_distance_to_next_disc)
UnboundLocalError: local variable 'approx_distance_to_next_disc' referenced before assignment
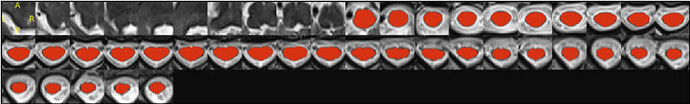
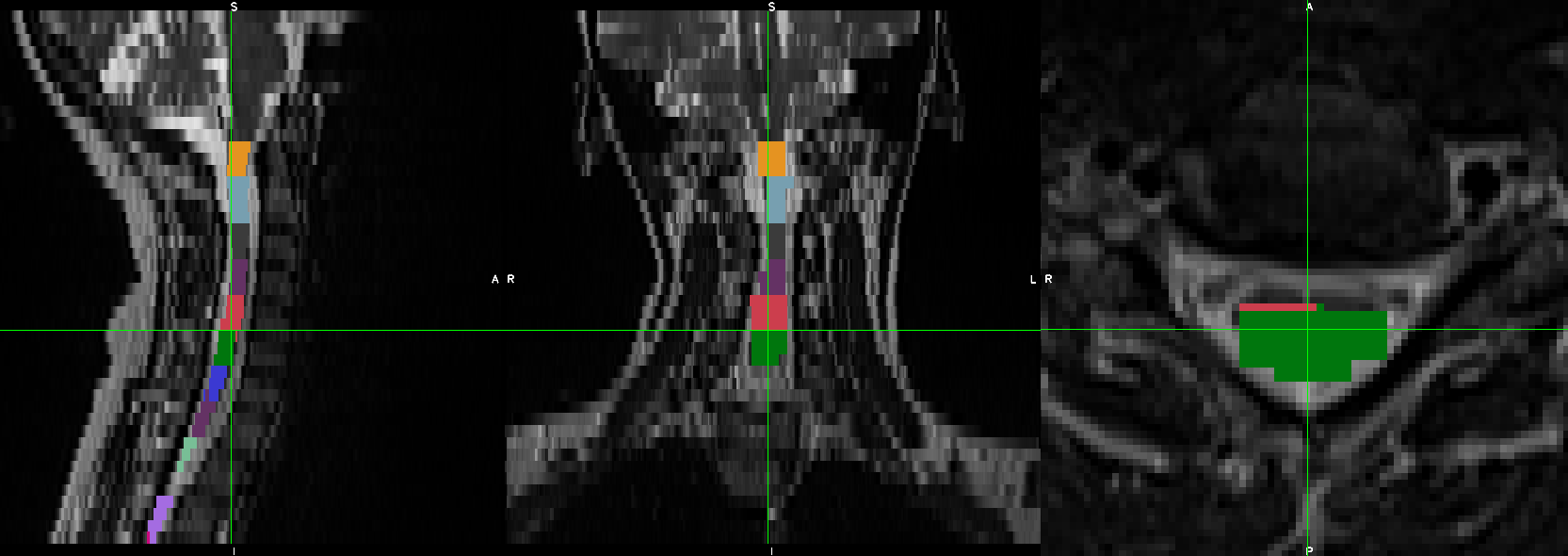
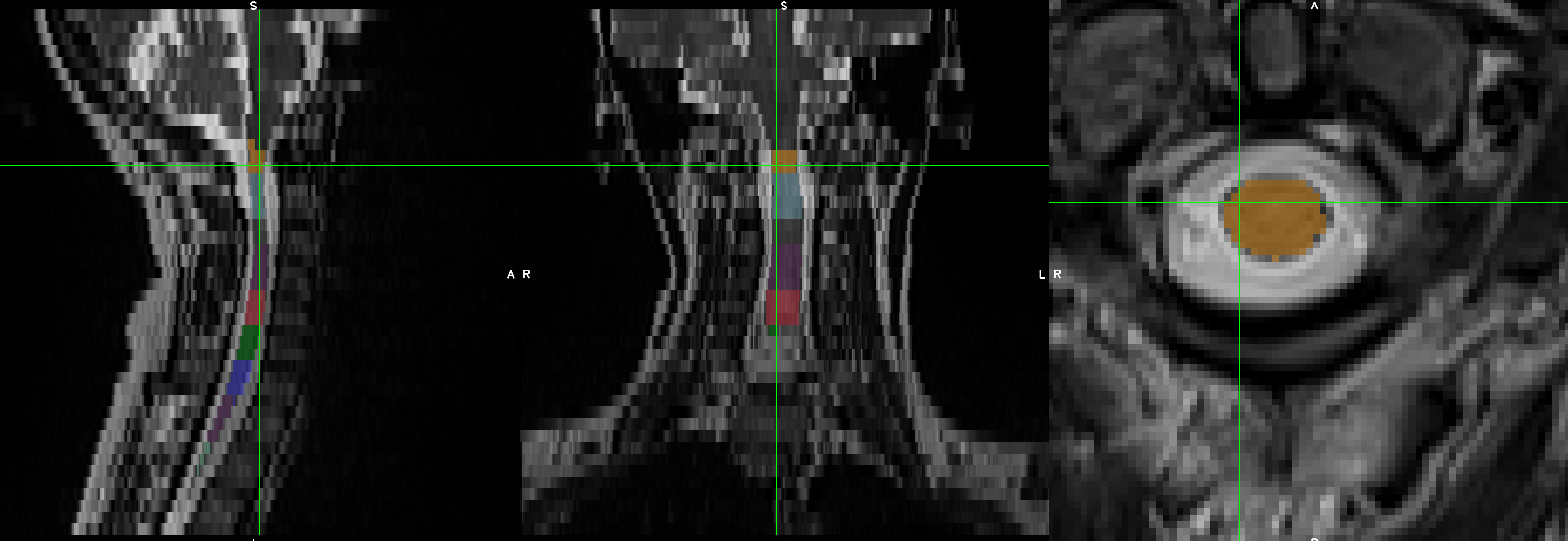
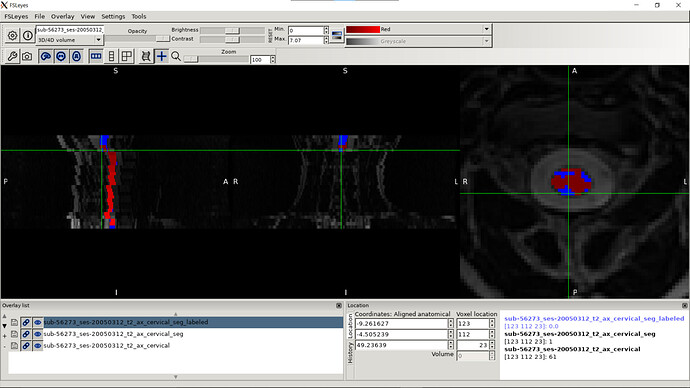
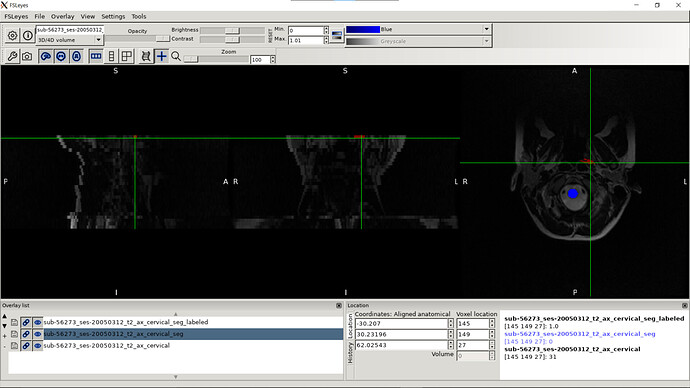
- When I was trying to run sct_register_multimodal it seems like it is in the right direction but the results still need some improvements:
sct_register_multimodal -i sag_cervical_seg_labeled.nii.gz -d ax_cervical.nii.gz -identity 1 -o ax_cervical_seg_labeled.nii.gz
I’ll appreciate your further help regarding those commands.
I’ll send you the original files by email.
Thanks a lot,
Juda