dear jcohenadad, I think you are busy , but if you have time , I wish you could help me .
because it is too hard for me .
waiting for your time. thank you very much.
I’ve looked at your diffusion MRI data. In order to register it with the PAM50 template, I would need to know what level (disc or mid-vertebral body) the volume is centered at.
Dear Jcohenadad. Thank you so much for your attention. This data is C5-C7.
Thank you.
Here is the script to process your data:
# Create variable name for convenience
file_dwi="TANG_WING_KUN_WIP_DTI_thk7_breath_1_5_1"
file_bvec=${file_dwi}.bvec
file_bval=${file_dwi}.bval
# Separate b=0 and DW images
sct_dmri_separate_b0_and_dwi -i ${file_dwi}.nii.gz -bvec ${file_bvec}
# Segment cord (1st pass -- just to get a rough centerline)
sct_propseg -i ${file_dwi}_dwi_mean.nii.gz -c dwi
# Create mask to help motion correction and for faster processing
sct_create_mask -i ${file_dwi}_dwi_mean.nii.gz -p centerline,${file_dwi}_dwi_mean_seg.nii.gz -size 30mm
# Crop data for faster processing
sct_crop_image -i ${file_dwi}.nii.gz -m mask_${file_dwi}_dwi_mean.nii.gz -o ${file_dwi}_crop.nii.gz
# Motion correction
sct_dmri_moco -i ${file_dwi}_crop.nii.gz -bvec ${file_dwi}.bvec -x spline
file_dwi=${file_dwi}_crop_moco
file_dwi_mean=${file_dwi}_dwi_mean
# Segment spinal cord (only if it does not exist)
sct_deepseg_sc -i ${file_dwi_mean}.nii.gz -c dwi -qc qc
file_dwi_seg=${file_dwi_mean}_seg
# Create labels, assuming that the mid-FOV is centered at C6 vertebral level
sct_label_utils -i ${file_dwi_seg}.nii.gz -create-seg -1,6 -o label_C6.nii.gz
# Register DWI data to the PAM50 template
sct_register_to_template -i ${file_dwi_mean}.nii.gz -s ${file_dwi_seg}.nii.gz -l label_C6.nii.gz -ref subject -c t1 -param step=1,type=seg,algo=centermassrot:step=2,type=im,algo=syn,metric=CC,slicewise=0,smooth=0,iter=3 -qc qc
# Rename warping field for clarity
mv warp_template2anat.nii.gz warp_template2dwi.nii.gz
mv warp_anat2template.nii.gz warp_dwi2template.nii.gz
# Warp template
sct_warp_template -d ${file_dwi_mean}.nii.gz -w warp_template2dwi.nii.gz -qc qc
# Compute DTI
sct_dmri_compute_dti -i ${file_dwi}.nii.gz -bvec ${file_bvec} -bval ${file_bval} -method standard
# Compute FA, MD and RD in WM and GM between C5 and C7 vertebral levels
sct_extract_metric -i dti_FA.nii.gz -f label/atlas -l 51,52 -vert 5:7 -o DWI_FA.csv -append 1
sct_extract_metric -i dti_MD.nii.gz -f label/atlas -l 51,52 -vert 5:7 -o DWI_MD.csv -append 1
sct_extract_metric -i dti_RD.nii.gz -f label/atlas -l 51,52 -vert 5:7 -o DWI_RD.csv -append 1
And here are the results (QC report and csv files): results.zip (382.8 KB)
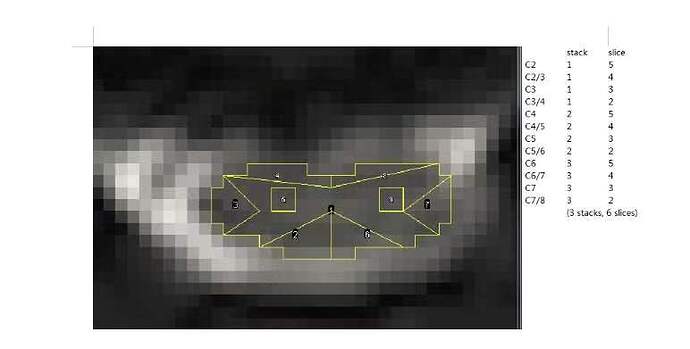
dear jcohenadad, I have a question . if the sct could get the ROI just like this? it is our team’s aim.
thank you . if you can, could you help me. I am sorry for bothering you a long time.
hi,
if you look at the usage of sct_extract_metric, you will notice that the labels can be called (via the flag -l). Here is a list of combined labels that you might find relevant:
50, spinal cord, 0:35
51, white matter, 0:29
52, gray matter, 30:35
53, dorsal columns, 0:3
54, lateral funiculi, 4:13
55, ventral funiculi, 14:29
and if you want, let’s say, to separate the right and left funiculi, you could edit the file info_label.txt (as instructed here) and add the following entries:
56, left funiculi, 4,6,8,10,12
57, right funiculi, 5,7,9,11,13
and then run sct_extract_metric with these new labels.
best,
Julien
dear jcohenadad , I have a new problem,in this script, "sct_label_utils -i mt1_seg.nii.gz -create-seg -1,4 -o label_c3c4.nii.gz ". How to modify is to label c3 instead of c3-c4 .I don’t know how to label the c3.
thank you .
This thread is getting very confusing. You are asking different questions based on changing datasets. Your post here refers to a DWI data, which covers C5 to C7. So, I wrote a script for analyzing this dataset.
Now, you are referring to a command which has nothing to do with the script I wrote, and apparently you are also referring to another dataset. This makes the whole conversation very confusing.
In order to move efficiently, I suggest you create a new thread for each specific question. Also, please, when you ask a question, please provide the context: what dataset (upload it if you can), what exactly would you like to quantify, what priors do you have (e.g., is the FOV centered at a specific level? if not, do you have another volume where discs are visible?).
Also, there is an upcoming SCT course in London next week, so I strongly suggest you attend it (will be live streamed and recorded).
Best,
Julien
Dear jcohenadad ,I am really sorry, because I am a student , I must follow the instructions of my teacher.
I have not understood the meaning of my teacher before.
But I think I have solved the most of the problems with your help. and there is one thing left.
Now, I think I should get the separate value of each C6.C6-C7.C7.C7-8. The course taught me to label the disc of C6-C7 and C7-8.but I could figure out how to label the C6 and C7.
I have a little knowledge of computer science , and my English is not good.if I did something wrong. Please forgive me.
By the way , I will listen to the course of London.thank you for your help.
The labeling is done for template registration.
The extraction of metrics uses vertebral levels (not disc levels) to extract metrics at specific vertebral levels.
So, if you want values at C3 mid-vertebral body, C3-C4 disc, C4 mid-vertebral body, etc. you could do the following:
sct_extract_metric -i dti_FA.nii.gz -f label/atlas -l 51,52 -vert 3:5 -perlevel 1 -o DWI_FA.csv
This will give you ONE value per mid-vertebral body level. Then, you average adjacent values (e.g. C3 and C4), which will give you the disc value (C3-C4).
dear jcohenadad , I have get the result of the dwi I want . but I think the fa value is much lower than the normal value. why this will happened?
could you check my code for a while ?
> file_dwi="TANG_WING_KUN_WIP_DTI_thk7_breath_1_5_1"
>
> file_bvec=${file_dwi}.bvec
>
> file_bval=${file_dwi}.bval
>
> Separate b=0 and DW images
>
> sct_dmri_separate_b0_and_dwi -i ${file_dwi}.nii.gz -bvec ${file_bvec}
>
> sct_propseg -i ${file_dwi}_dwi_mean.nii.gz -c dwi
>
> sct_create_mask -i ${file_dwi}_dwi_mean.nii.gz -p centerline,${file_dwi}_dwi_mean_seg.nii.gz -size 30mm
>
> sct_crop_image -i ${file_dwi}.nii.gz -m mask_${file_dwi}_dwi_mean.nii.gz -o ${file_dwi}_crop.nii.gz
>
> sct_dmri_moco -i ${file_dwi}_crop.nii.gz -bvec ${file_dwi}.bvec -x spline
>
> file_dwi=${file_dwi}_crop_moco
>
> file_dwi_mean=${file_dwi}_dwi_mean
>
> # # Segment spinal cord (only if it does not exist)
>
> sct_deepseg_sc -i ${file_dwi_mean}.nii.gz -c dwi -qc qc
>
> file_dwi_seg=${file_dwi_mean}_seg
>
> # # # Create labels, assuming that the mid-FOV is centered at C6 vertebral level
>
> # sct_label_utils -i ${file_dwi_seg}.nii.gz -create-seg 6,8 -o label_C6.nii.gz
>
> sct_label_utils -i ${file_dwi_seg}.nii.gz -vert-body 6,8 -o label_C6.nii.gz
>
> sct_register_to_template -i ${file_dwi_mean}.nii.gz -s ${file_dwi_seg}.nii.gz -l label_C6.nii.gz -ref subject -c t1 -param step=1,type=seg,algo=centermassrot:step=2,type=im,algo=syn,metric=CC,slicewise=0,smooth=0,iter=3 -qc qc
>
> mv warp_template2anat.nii.gz warp_template2dwi.nii.gz
>
> mv warp_anat2template.nii.gz warp_dwi2template.nii.gz
>
> sct_warp_template -d ${file_dwi_mean}.nii.gz -w warp_template2dwi.nii.gz -qc qc
>
> sct_dmri_compute_dti -i ${file_dwi}.nii.gz -bvec ${file_bvec} -bval ${file_bval} -method standard
>
> sct_maths -i label/atlas/PAM50_atlas_00.nii.gz -add $(for i in `seq -w 2 2 36`; do echo "label/atlas/PAM50_atlas_${i}.nii.gz"; done) -o label/atlas/PAM50_atlas_left.nii.gz
>
> sct_maths -i label/atlas/PAM50_atlas_01.nii.gz -add $(for i in `seq -w 3 2 35`; do echo "label/atlas/PAM50_atlas_${i}.nii.gz"; done) -o label/atlas/PAM50_atlas_right.nii.gz
>
> # sct_extract_metric -i dti_FA.nii.gz -f label/atlas -l 51,52 -vert 3:5 -perlevel 1 -o DWI_FA.csv
>
> sct_extract_metric -i label/atlas/PAM50_atlas_left.nii.gz -f label/atlas -l 52,53,54,55 -vert 6:8 -perlevel 1 -o DWI_FA.csv -append 1
>
> sct_extract_metric -i label/atlas/PAM50_atlas_right.nii.gz -f label/atlas -l 52,53,54,55 -vert 6:8 -perlevel 1 -o DWI_FA.csv -append 1
>
> # sct_extract_metric -i label/atlas/PAM50_atlas_left.nii.gz -f label/atlas -l 52,53,54,55 -vert 6:8 -o DWI_FA.csv -append 1
>
> # sct_extract_metric -i label/atlas/PAM50_atlas_right.nii.gz -f label/atlas -l 52,53,54,55 -vert 6:8 -o DWI_FA.csv -append 1
Two issues.
Issue 1:
with this line:
> sct_maths -i label/atlas/PAM50_atlas_00.nii.gz -add $(for i in `seq -w 2 2 36`; do echo "label/atlas/PAM50_atlas_${i}.nii.gz"; done) -o label/atlas/PAM50_atlas_left.nii.gz
you are including the CSF in your mask, so it is expected that FA would be abnormally low.
instead of using this approach of creating a mask by adding labels, i suggest you stick to my instructions here, i.e., ONLY change the file info_label.txt by adding the tracts you like.
Issue 2:
with this line:
> sct_extract_metric -i label/atlas/PAM50_atlas_left.nii.gz -f label/atlas -l 52,53,54,55 -vert 6:8 -perlevel 1 -o DWI_FA.csv -append 1
you are quantifying the value of a binary mask (instead of the FA).
Dear jcohenadad.
Thank you for your reply, but could you help me to change the code ?I am afraid I do it wrong again!
Thank you!
dear jcohenadad
with this line:
> sct_extract_metric -i label/atlas/PAM50_atlas_left.nii.gz -f label/atlas -l 52,53,54,55 -vert 6:8 -perlevel 1 -o DWI_FA.csv -append 1
you are quantifying the value of a binary mask (instead of the FA).
I don’t know how to change this line. it is a little difficult for me.
I don’t know how to change this line. it is a little difficult for me.
You should not use that line. Just modify the file info_label.txt by adding those two lines:
56, left funiculi, 4,6,8,10,12
57, right funiculi, 5,7,9,11,13
For information, here is the modified file: info_label.txt (2.8 KB)
Copy the file under your SCT installation folder (let’s call it $SCT_DIR): $SCT_DIR/data/PAM50/atlas/ (replace the existing one)
Then re-run this code, but replace the last lines (everything below “# Compute FA, MD and RD in WM…”) with:
sct_extract_metric -i dti_FA.nii.gz -f label/atlas -l 51,52,56,57 -vert 5:7 -o DWI_FA.csv -append 1
sct_extract_metric -i dti_MD.nii.gz -f label/atlas -l 51,52,56,57 -vert 5:7 -o DWI_MD.csv -append 1
sct_extract_metric -i dti_RD.nii.gz -f label/atlas -l 51,52,56,57 -vert 5:7 -o DWI_RD.csv -append 1
Dear jcohenadad .
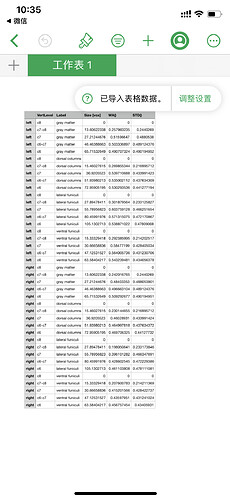
Your code will result a outcome like this.
But I want a result like this
Not just the left and right result. But also a result including left dorsal , ventral ,and lateral.and right dorsal ,ventral, and lateral.
Could you help me?
dear jcohenadad , maybe I expressed myself not clearly. could you tell me where is my mistake ?
Dear jcohenadad,
could you provide the help to me for this problem?
Thank you.
Hi
You can use other labels and specify them with the flag -l
To see the list of labels available run: sct_extract_metric