Hello,
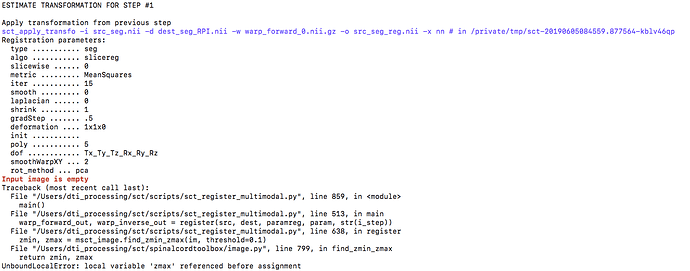
I am attempting to register a C2-T2 14 slice b0 DTI scan to its mFFE using the sct_register_multimodal tool. However, I am receiving the attached error that says that zmax is referenced before assignment and that the input image is empty. I am using the following command for the registration: `
sudo sct_register_multimodal -i dti_crop_moco_b0_mean.nii -d ~/Documents/Patient_Data_Folder/“$dirName”/DTI_Data/niftis/mFFE_crop.nii -o b02mFFE.nii.gz -owarp b02mFFE_warp.nii.gz -iseg dti_crop_moco_dwi_mean_seg_copy.nii.gz -dseg ~/Documents/Patient_Data_Folder/“$dirName”/DTI_Data/niftis/mFFE_crop_seg.nii.gz -param step=1,type=seg,algo=slicereg,metric=MeanSquares,gradStep=.5,iter=15:step=2,type=seg,algo=affine,metric=MeanSquares,gradStep=.05,iter=15, shrink=1:step=3,type=im,algo=syn,metric=MI,iter=15,shrink=1 -qc ~/Documents/Patient_Data_Folder/“$dirName”/Raw_Data/NIIX_Conv/DTI_Processing
The DTI and mFFE segmentation files used as the input image in the first parameter step have segmentation data and last for 14 slices. The respective mFFE and DTI images are similarly non-empty. I would appreciate some more clarification on the error and possible steps to remedy it. Appreciate your help.
Regards,
Ashwin
`