Hi Rosella,
However, sct_register_to_template from 3dT1 to PAM50 and viceversa produced an error , as already mentioned in previous discussions on the forum.
I see you had issues with sct_label_vertebrae here, but I cannot find the specific issue you encountered with sct_register_to_template. Which issue are you referring to?
As regards registration to the template, the problem I had was that the registration output I obtained included some CSF at some SC level due to size differences between PAM 50 ADULT atlas and my NEONATE images. So, in order to be sure to exclude CSF, I reduced PAM50 labels corresponding to the WM tract of interest (CST in this case) to just their intersection with SC segmentation of my images.
I used sct to compute DTI maps and DIPY to compute DKI maps.
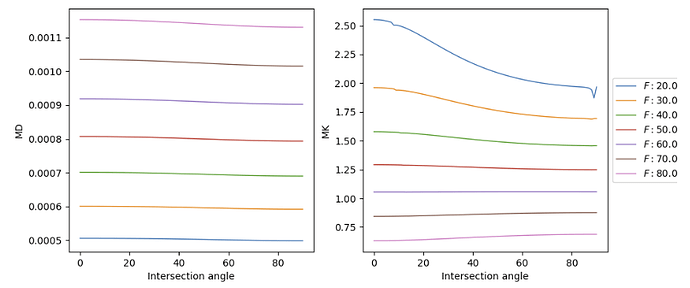
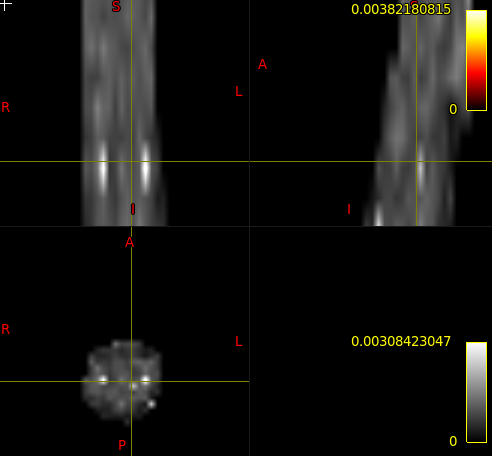
Here you find attached the script with the processing steps ( script_kurtosis.sh ),the python DIPY script ( dipy_DKI.py ), all the images produced through applying the script, the DTI and DKI maps, and the average metrics results over lateral CST.
Do you think this is the resolutive approach? Could it work with other WM tracts?
A couple of comments:
- Why are you outputting the CSF from the propseg segmentation?
- Your script has an error at line 12:
-ofolderis missing an argument. - No, it is not a good idea to reduce the intersection between the registered PAM50 data (e.g. WM) and your cord segmentation, because (i) if there is a misregistration, simply taking the intersect will not solve the other issues (e.g. misregistration inside the cord) and (ii) modifying the partial volume information will violate the assumption made while running
sct_extract_metricusing maximum likelihood or maximum a posteriori estimation. The right thing to do is change the registration parameter to allow more degree of freedom, which is what i did in the updated script. - The data you shared in your previous post do not include the file
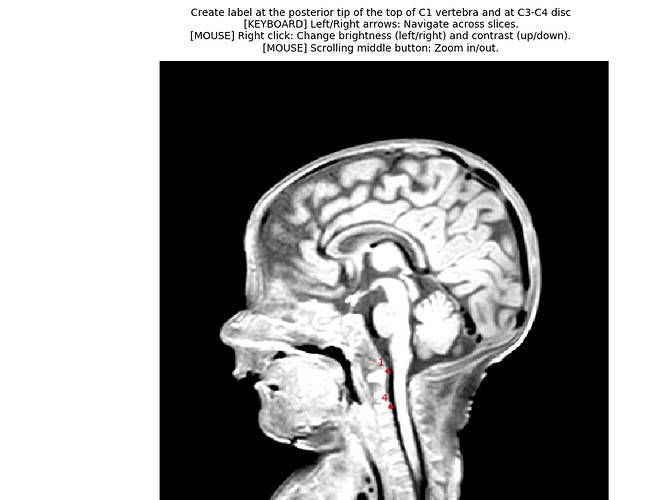
3dT1.nii, therefore I cannot get the labels from this file. So instead, I assumed realistic vertebral levels for two slices on the kurtosis image directly, and I used that to generate two labels. You will need to update those numbers according to the proper levels. - I recommend you use the QC feature. It help validating the quality of segmentation, registration to template, etc. and also to share your analysis and keep that as a record of what you actually ran (because the exact syntax is included in the QC report, along with SCT version).
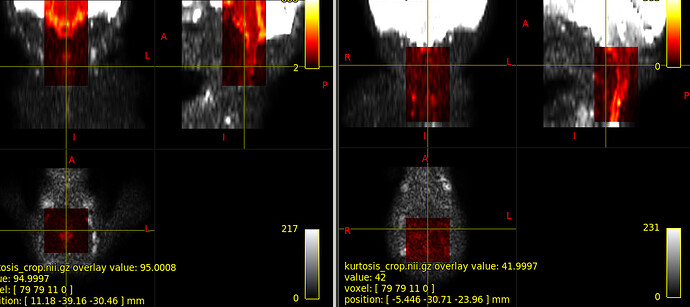
So, I modified the script to address all those issues: kurtosis_JCA.sh (2.7 KB) . Here are the QC report and DTI results so you can make sure you can reproduce my results: qc-dti.zip (428.6 KB)
I cannot clearly see if registration of the atlas to the template is fine also longitudinally, with a perfect correspondence along spinal levels, due to different sizes.
Please note that your workflow is using vertebral levels as references for alignment with the PAM50 template, not spinal levels.
Best,
Julien