Dear all,
I am trying to calculate CSA in the upper area of the cervical spine which is covered within Brain MRI data.
It would be sufficient to calculate results from sct_deepseg_sc
To compare, it would be even better to limit CSA till end of C4 but I struggle with that application.
Please advise
Thx
Hi @mvaro2002,
Have you see this post?
Cheers,
Julien
Thank you for the quick reply,
just to elaborate:
I have T1/MRAGE & T2 FLAIR Images
I converted them from DICOM to NIFTI using MRIcroGL
Running sct_deepseg_sc I get this error:
Spinal cord was not detected using “-centerline cnn”. Please try another “-centerline” method.
I tried another scan and this error occurred:
Spinal Cord Toolbox (4.2.2)
Config deepseg_sc:
Centerline algorithm: cnn
Brain in image: True
Kernel dimension: 2d
Contrast: t1
Threshold: 0.15
Create temporary folder (/var/folders/09/_0t8bgrs6qggzpz96846h1480000gn/T/sct-20200414161117.346703-pbdo95ln)…
Reorient the image to RPI, if necessary…
Finding the spinal cord centerline…
WARNING: To avoid intensity overflow due to convertion to uint8, intensity will be rescaled to the maximum quantization scale.
Brain section detected.
Remove temporary files…
rm -rf /var/folders/09/_0t8bgrs6qggzpz96846h1480000gn/T/sct-20200414161124.223477-dpa745y9
Cropping brain section.
Cropping the image around the spinal cord…
Normalizing the intensity…
Segmenting the spinal cord using deep learning on 2D patches…
Reassembling the image…
Resampling the segmentation to the native image resolution using linear interpolation…
Binarizing the resampled segmentation…
Traceback (most recent call last):
File “/Users/XXX/sct_4.2.2/scripts/sct_deepseg_sc.py”, line 208, in
main()
File “/Users/XXX/sct_4.2.2/scripts/sct_deepseg_sc.py”, line 192, in main
threshold_seg=threshold, remove_temp_files=remove_temp_files, verbose=verbose)
File “/Users/XXX/sct_4.2.2/spinalcordtoolbox/deepseg_sc/core.py”, line 554, in deep_segmentation_spinalcord
im_seg_r_postproc = post_processing_volume_wise(im_seg_r)
File “/Users/XXX/sct_4.2.2/spinalcordtoolbox/deepseg_sc/postprocessing.py”, line 144, in post_processing_volume_wise
im_seg = _remove_isolated_voxels_on_the_edge(im_seg)
File “/Users/XXX/sct_4.2.2/spinalcordtoolbox/deepseg_sc/postprocessing.py”, line 110, in _remove_isolated_voxels_on_the_edge
metrics, _ = compute_shape(im_seg, angle_correction=False)
File “/Users/XXX/sct_4.2.2/spinalcordtoolbox/process_seg.py”, line 57, in compute_shape
min_z_index, max_z_index = min(Z), max(Z)
ValueError: min() arg is an empty sequence
would you mind sending me these two images so I can try to see what’s the problem?
If you prefer to send them to me privately you can use: “jcohen [at] polymtl.ca”
Cheers
Julien
Hi @mvaro2002,
I was not able to reproduce your errors. The segmentation worked fine in both datasets. Here is the QC report, which includes the syntax I used: qc_20200414215530.zip (967.4 KB)
By the way: I noticed that both of your scans were not centred in the medial plane and were missing a large chunk of the brain, which is quite uncommon for brain T1w scans.
Best,
Julien
Hi @mvaro2002,
Just following up on this issue. Has it been solved on your end?
Cheers
Julien
Hi @mvaro2002,
following our email conversation, below is the code that I used to compute CSA between two time points:
#!/bin/bash
FILES=(Test_20180830_T1 Test_20180823_T1)
for file in ${FILES[@]}; do
# Segment cord
sct_deepseg_sc -i ${file}.nii.gz -c t1 -qc qc
# Label vertebrae
sct_label_vertebrae -i ${file}.nii.gz -s ${file}_seg.nii.gz -c t1 -qc qc
# Compute average CSA between C1-C3 level
# Note: in your case, both scans show the full C3 while C4 is inconsistently cut,
# so I recommend discarding C4 for CSA computation.
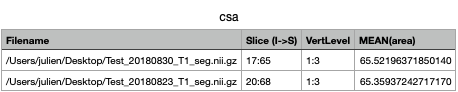
sct_process_segmentation -i ${file}_seg.nii.gz -vertfile ${file}_seg_labeled.nii.gz -vert 1:3 -o csa.csv -append 1
done
You can download the script here: script_mihael.sh (547 Bytes)
And here is the results: csa.csv (1.2 KB)
As you can see, CSA values are reasonably similar:
Cheers,
Julien
1 Like
