The following example will show how to compute CSA on T1w scans which were acquired for the brain, but where the upper cervical spinal cord is visible:
# This code has been tested with SCT v4.2.1
# Download example data
sct_download_data -d sct_example_data
# Crop to simulate a brain-only scan with the cord visible down to C2-C3 disc
sct_crop_image -i t1.nii.gz -zmin 125
# Segment the spinal cord, specifying that a brain is present in the image
sct_deepseg_sc -i t1_crop.nii.gz -c t1 -brain 1 -centerline cnn
# Label vertebrae
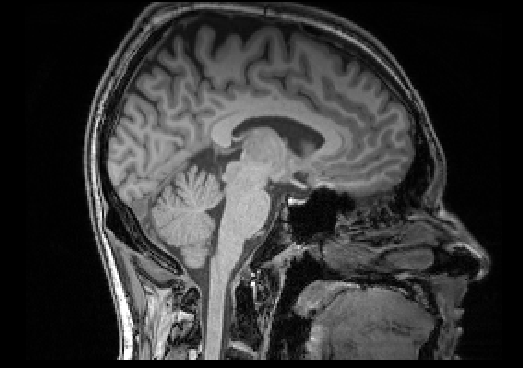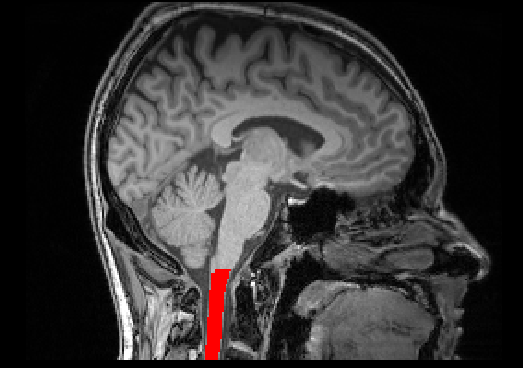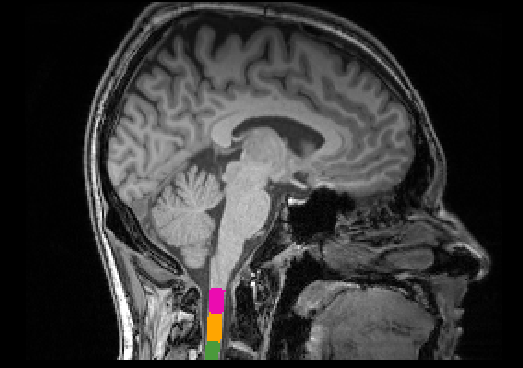
sct_label_vertebrae -i t1_crop.nii.gz -s t1_crop_seg.nii.gz -c t1
# Compute average CSA between C1-C2 level
sct_process_segmentation -i t1_crop_seg.nii.gz -vertfile t1_crop_seg_labeled.nii.gz -vert 1:2
Here is the output CSA (including other morphometric measures): csa.csv (777 Bytes)