Dear forum
I have run into a problem using sct_register_multimodal, sct version 4.0.0 beta 2. Or not necessarily a problem, just something I don’t understand and that might be worth highlighting.
I am registering an MT weighted scan to a MEDIC. Resolutions are 625um^2 and 450um^2 respectively. The MEDIC was a 2D scan, the MT a 3D one.
When I use a non-rigid algorithm (syn) to coregister, the spinal cord in the resulting registered MTw scan appears compressed, and the vertebrate are comparatively enlarged. Please see the picture for more info.
MEDIC:
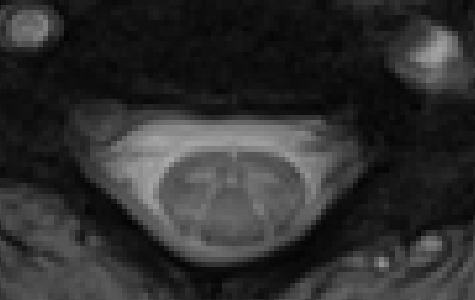
Centermass coreg:
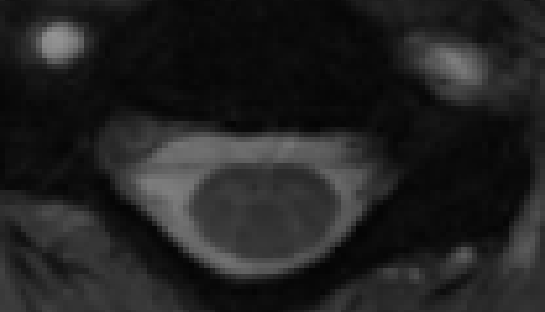
Syn coreg:
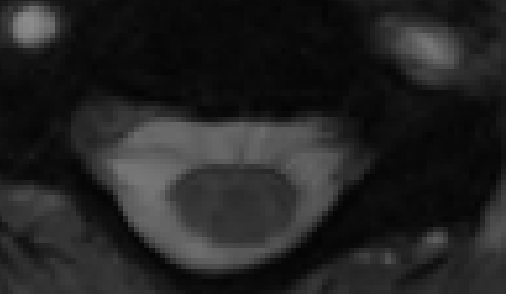
I tried supplying a mask (35mm, cylinder), it did not improve the situation. I am getting this issue when I use non-linear transformations, but not for linear ones or center-of-mass ones. Why is this happening? What can I do to avoid this issue? Is this WAD?
Data here: Dropbox - 4JCA.zip - Simplify your life
Password: spinalcord
Commands for each output, and wether the issue appears:
No issue on these:
sct_register_multimodal -i MTw_mean.nii -iseg MTw_mean_seg.nii -d MEDIC.nii -dseg MEDIC_seg.nii -param step=1,algo=translation,type=im -m mask_MEDIC.nii -o ./Examples/translation.nii
sct_register_multimodal -i MTw_mean.nii -iseg MTw_mean_seg.nii -d MEDIC.nii -dseg MEDIC_seg.nii -param step=1,algo=rigid,type=im -m mask_MEDIC.nii -o ./Examples/rigid.nii
sct_register_multimodal -i MTw_mean.nii -iseg MTw_mean_seg.nii -d MEDIC.nii -dseg MEDIC_seg.nii -param step=1,algo=affine,type=im -m mask_MEDIC.nii -o ./Examples/affine.nii
sct_register_multimodal -i MTw_mean.nii -iseg MTw_mean_seg.nii -d MEDIC.nii -dseg MEDIC_seg.nii -param step=1,algo=slicereg,type=im -m mask_MEDIC.nii -o ./Examples/slicereg.nii
sct_register_multimodal -i MTw_mean.nii -iseg MTw_mean_seg.nii -d MEDIC.nii -dseg MEDIC_seg.nii -param step=1,algo=centermass,type=seg -m mask_MEDIC.nii -o ./Examples/centermass.nii
sct_register_multimodal -i MTw_mean.nii -iseg MTw_mean_seg.nii -d MEDIC.nii -dseg MEDIC_seg.nii -param step=1,algo=centermassrot,type=seg -m mask_MEDIC.nii -o ./Examples/centermassrot.nii
Issue present here:
sct_register_multimodal -i MTw_mean.nii -iseg MTw_mean_seg.nii -d MEDIC.nii -dseg MEDIC_seg.nii -param step=1,algo=syn,type=im -m mask_MEDIC.nii -o ./Examples/syn.nii
Slicereg does not change the outcome
sct_register_multimodal -i MTw_mean.nii -iseg MTw_mean_seg.nii -d MEDIC.nii -dseg MEDIC_seg.nii -param step=1,algo=syn,type=im,slicewise=1 -m mask_MEDIC.nii -o ./Examples/syn_slicewise.nii
I tried running it with bsplinesyn, but it did not finish in 2 hours.
Different artefact here
sct_register_multimodal -i MTw_mean.nii -iseg MTw_mean_seg.nii -d MEDIC.nii -dseg MEDIC_seg.nii -param step=1,algo=columnwise,type=seg -m mask_MEDIC.nii -o ./Examples/colwise.nii
Best wishes
Daniel