Hi Julien,
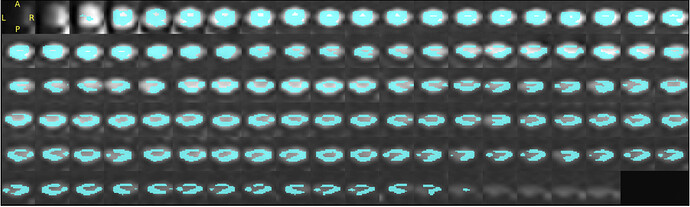
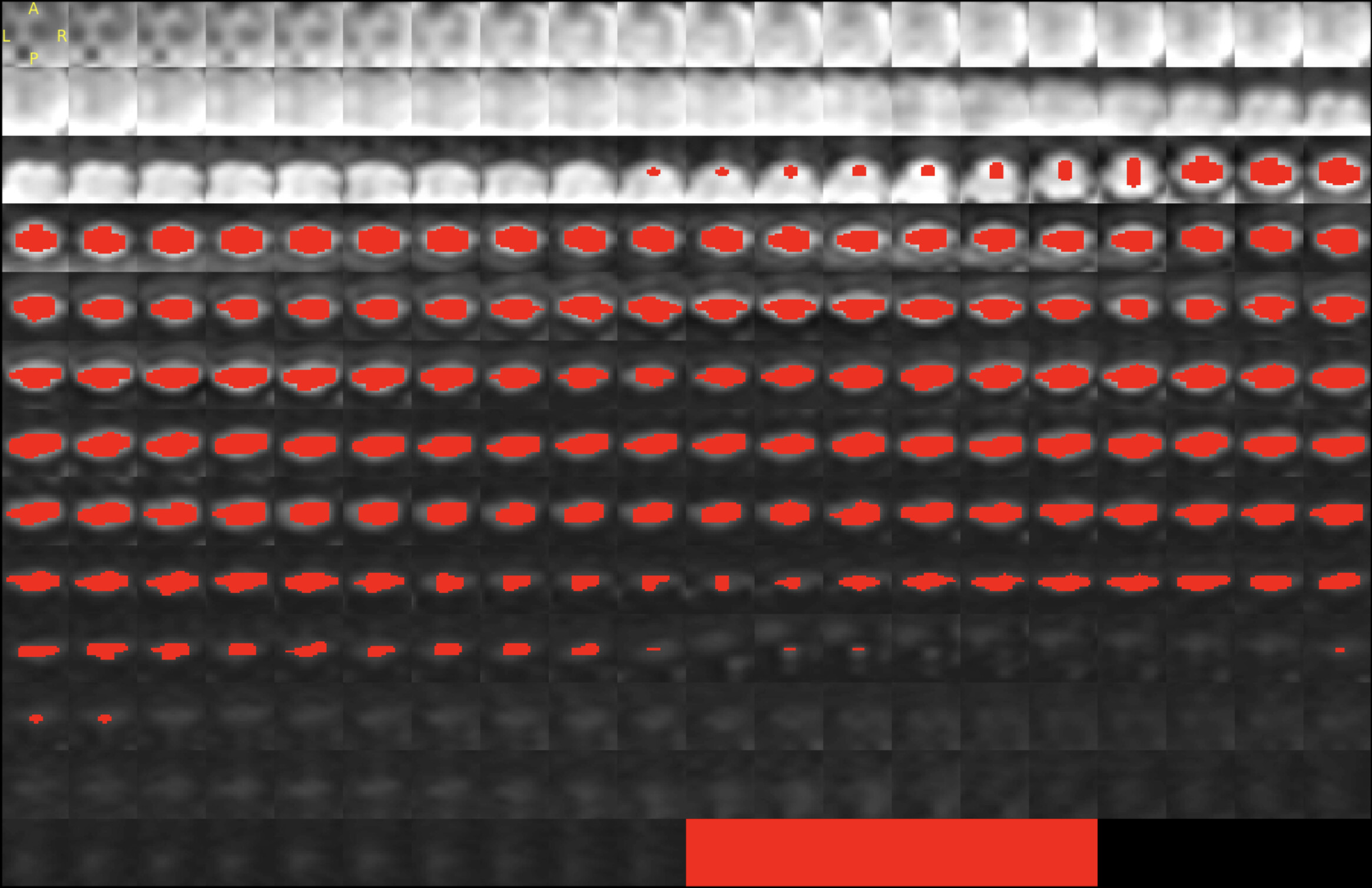
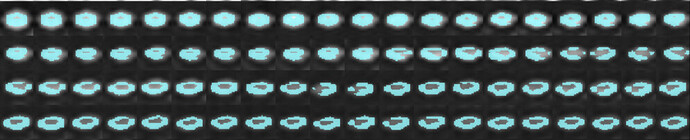
I am working on spinal cord DTI images acquired as sagittal images. I find the registration to PAM50 template is not as good as expected (attached are the outputs)Can you let me know how I can improve the registration?
How can I send you the data- I can’t seem to be able to upload the DTI scan?
Thanks,
Aditya
SCRIPT:
sct_propseg -i t2.nii.gz -c t2 -qc ~/qc_DTI_Cordotomy
sct_deepseg_sc -i t2.nii.gz -c t1 -ofolder deepseg -qc ~/qc_DTI_Cordotomy
———————
sct_label_vertebrae -i t2.nii.gz -s t2_seg.nii.gz -c t2 -qc ~/qc_DTI_Cordotomy
# If automatic labeling did not work, you can initialize with manual identification of C2-C3 disc:
sct_label_utils -i t2.nii.gz -create-viewer 3 -o label_c2c3.nii.gz -msg "Click at the posterior tip of C2/C3 inter-vertebral disc"
sct_label_vertebrae -i t2.nii.gz -s t2_seg.nii.gz -c t2 -initlabel label_c2c3.nii.gz -qc ~/qc_DTI_Cordotomy
—————————————
sct_label_utils -i t2_seg_labeled.nii.gz -vert-body 3,9 -o t2_labels_vert.nii.gz
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -l t2_labels_vert.nii.gz -c t2 -qc ~/qc_DTI_Cordotomy
sct_warp_template -d t2.nii.gz -w warp_template2anat.nii.gz -qc ~/qc_DTI_Cordotomy
——————————————
# Compute mean dMRI from dMRI data
sct_maths -i dmri.nii.gz -mean t -o dmri_mean.nii.gz
# Segment SC on mean dMRI data
# Note: This segmentation does not need to be accurate-- it is only used to create a mask around the cord
sct_propseg -i dmri_mean.nii.gz -c dwi -qc ~/qc_DTI_Cordotomy
# Create mask (for subsequent cropping)
sct_create_mask -i dmri_mean.nii.gz -p centerline,dmri_mean_seg.nii.gz -size 35mm
# Crop data for faster processing
sct_crop_image -i dmri.nii.gz -m mask_dmri_mean.nii.gz -o dmri_crop.nii.gz
# Motion correction (moco)
sct_dmri_moco -i dmri_crop.nii.gz -bvec bvecs.txt
# Segment SC on motion-corrected mean dwi data (check results in the QC report)
sct_propseg -i dmri_crop_moco_dwi_mean.nii.gz -c dwi -qc ~/qc_DTI_Cordotomy
# Register template->dwi via t2s to account for GM segmentation
sct_register_multimodal -i dmri_crop_moco_b0_mean.nii.gz -iseg dmri_crop_moco_dwi_mean_seg.nii.gz -d t2.nii.gz -dseg t2_seg.nii.gz -param step=1,type=seg,algo=slicereg,smooth=5:step=2,type=im,algo=syn,metric=CC,smooth=0,slicewise=0,iter=3 -qc ~/qc_DTI_Cordotomy
# Rename warping fields for clarity
mv warp_PAM50_t12dmri_crop_moco_dwi_mean.nii.gz warp_template2dmri.nii.gz
mv warp_dmri_crop_moco_dwi_mean2PAM50_t1.nii.gz warp_dmri2template.nii.gz
# Warp template
sct_warp_template -d dmri_crop_moco_dwi_mean.nii.gz -w warp_template2dmri.nii.gz -qc ~/qc_DTI_Cordotomy
# Check results in the QC report