Hi @jcohenadad and @Charley_Gros,
please find attached screenshots results for the new model…
Command used here is available in the latest version SCT v5.2.0
It was used as instructed by @Charley_Gros
sct_deepseg -install-task seg_mice_gm-wm_dwi
… and can be used as follows:
sct_deepseg -i NIFTI_IMAGE -task seg_mice_gm-wm_dwi
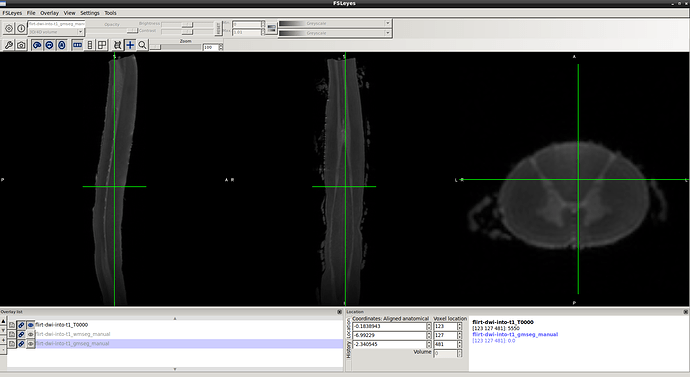
1- dwi data before processing…
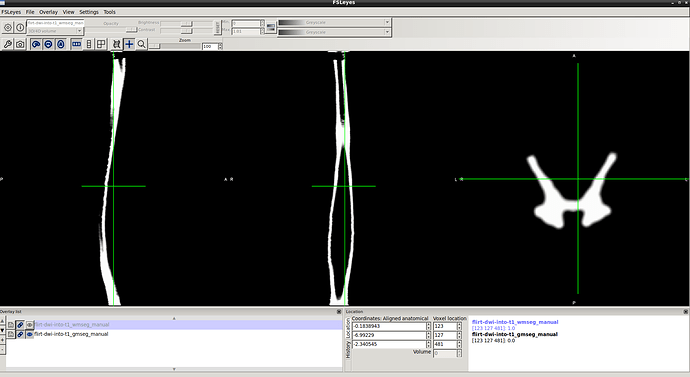
2- gm seg result …
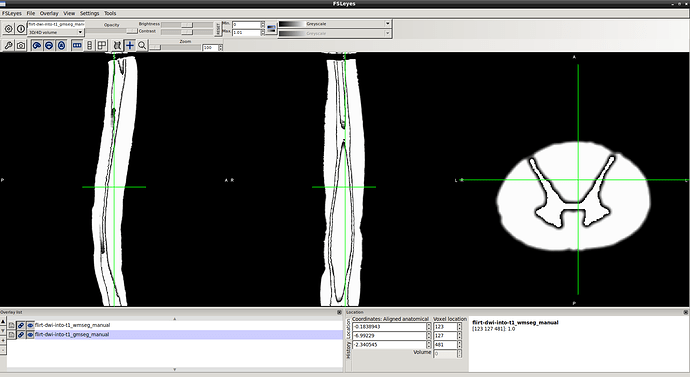
3- wm-gm seg result…
Many many thanks for this robust model and thank you for all SCT team for continues help and support to the community…
Cheers,
Ibrahim
1 Like
Fantastic!!!  Thank you so much @Charley_Gros for these efforts, and @ihattan for the feedback, it is greatly appreciated.
Thank you so much @Charley_Gros for these efforts, and @ihattan for the feedback, it is greatly appreciated.
1 Like
I would like to clarify something on this thread: it all started with @ihattan not being able to analyse human ex vivo data (see: Error when running sct_get_centerline).
Then, @Charley_Gros trained a new deepseg model, called seg_mice_gm-wm_dwi (see: Error when running sct_get_centerline). So: this model was trained on mouse data (assumed from the name of the model). @ihattan can you please confirm that the data you sent to Charley were indeed mouse data and not human data?
If so, what is the rationale for segmenting human ex vivo data using a mouse ex vivo model?
Hi @jcohenadad & @Charley_Gros,
Thank you very much @jcohenadad for getting back with these valuable information. The data was for DW human exvivo not mouse. There was just a mistake when named the model for a mice instead human I think. Please note that the T1 ex vivo template was generated for T1 from the same samples.
I hope this clarify your enquiry.
Many many thanks in advance for your effort and time to solve these issues.
Cheers,
Ibrahim
Thank you for the quick response Ibrahim,
OK, it all makes sense now. I’ve further confirmed it by looking at the dimensions of the data (see here). We will fix the name of the model then.
@ihattan since we’re at it: what is the contrast of the image that was used to train the model? You mention DW data, but DW acquisition is 4D (ie: it includes DW files and b=0 files which are more T2w). Was the b=0 image used? (it should have, because it is the one that shows best gray/white matter contrast).
I did the ground truth segmentation for wm on b0 image. @Charley_Gros which contrast was used to train the model?
could you please provide more details for @jcohenadad ?
Many thanks @jcohenadad & @Charley_Gros for your time…
Cheers,
Ibrahim
so it is very likely that Charley used the b=0 image then. So I would not call it a “DW model” but rather a T2 model.
Yes correct, I confirm: I used the b=0 image.
1 Like
 Thank you so much
Thank you so much