Dear SCT Development Team,
I hope this email finds you well.
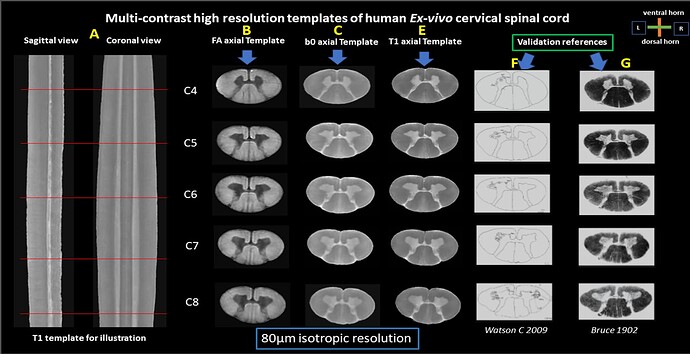
I have generated high-resolution multi-contrast templates of the ex vivo cervical spinal cord at 80-micron isotropic resolution using antsMultivariateTemplateConstruction2.sh script. These templates include b0, T1-weighted, and Fractional Anisotropy (FA) contrasts, covering spinal cord segments C4 to C8 as seen below.
1. Registration to PAM50 Atlas
I intend to register these templates to the PAM50 atlas for region-of-interest (ROI) analyses. While the SCT documentation and courses primarily focus on in vivo registration pipelines, no ex vivo-specific workflow has been provided.
Previously, I attempted to use the in vivo registration pipeline detailed in SCT, but I encountered challenges when applying it to the native space and these high-resolution templates. Unfortunately, due to a recent workstation update, I lost the results from that initial analysis. If you believe that approach could still be effective, I am happy to reanalyze and share the steps and results.
Alternatively, could you recommend a more robust or customized pipeline optimized for high-resolution ex vivo datasets?
2. Segmentation of Spinal Cord and Gray Matter
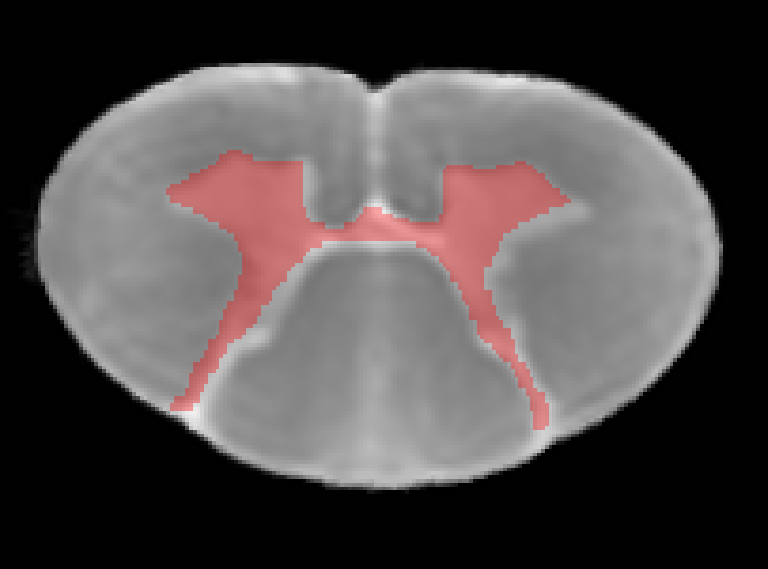
To enable accurate registration, I also aim to perform both spinal cord and gray matter (GM) segmentations. When applying the existing SCT-trained model for reference (Error when running sct_get_centerline - #20 by ihattan)) to the b0 template segmentation, I encountered issues with achieving accurate GM segmentation. Below is a screenshot showing the axial b0 template output for reference.
syntax I used for this segmentation:
sct_deepseg -i b0_template.nii.gz -task seg_exvivo_gm-wm_t2
Could you please provide guidance on optimizing this process, such as adjusting model parameters, modifying the syntax, or exploring alternative methods specifically suited for ex vivo data segmentation?
If it would be helpful, I am more than willing to share the data and results for further evaluation and potential optimization of the registration pipeline.
Your invaluable guidance has played a crucial role in the progress of this project, and I sincerely appreciate your continued support.
I am excited about the upcoming SCT course, and I believe that incorporating a dedicated session on ex vivo processing, labeling, and registration to the PAM50 template/atlas would be highly beneficial, particularly with the availability of high-resolution data.
Best regards,
Ibrahim