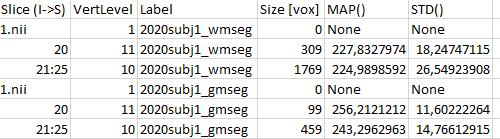
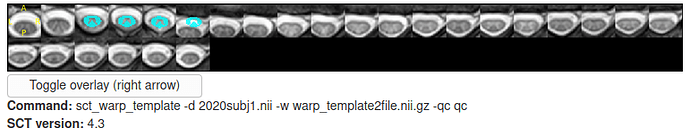
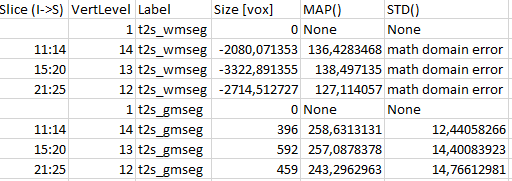
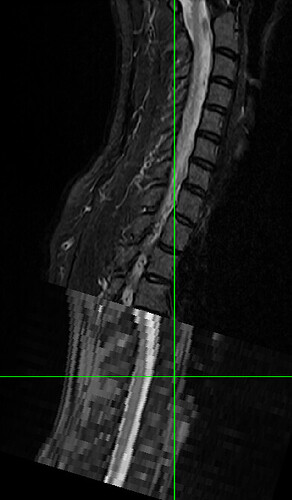
Here is the second half:
Move warping fields...
cp step2Warp.nii.gz /tmp/sct-20201029153546.541065-register-ocgl30mg
cp step2InverseWarp.nii.gz /tmp/sct-20201029153546.541065-register-ocgl30mg
rm -rf /tmp/sct-20201029154556.837548-register-tzn1pfg9
Concatenate transformations...
Parse list of warping fields...
Check file existence...
OK: dest.nii
OK: /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/analysisFiles/warp_template2anat.nii.gz
OK: warp_forward_1.mat
OK: warp_forward_2.nii.gz
/home/nirims/sct_4.3/bin/isct_ComposeMultiTransform 3 warp_final.nii.gz -R dest.nii warp_forward_2.nii.gz warp_forward_1.mat /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/analysisFiles/warp_template2anat.nii.gz # in /tmp/sct-20201029153546.541065-register-ocgl30mg
Generate output files...
File created: warp_src2dest.nii.gz
Parse list of warping fields...
Check file existence...
OK: src.nii
OK: warp_inverse_2.nii.gz
OK: warp_forward_1.mat
OK: /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/analysisFiles/warp_anat2template.nii.gz
/home/nirims/sct_4.3/bin/isct_ComposeMultiTransform 3 warp_final.nii.gz -R src.nii /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/analysisFiles/warp_anat2template.nii.gz -i warp_forward_1.mat warp_inverse_2.nii.gz # in /tmp/sct-20201029153546.541065-register-ocgl30mg
Generate output files...
File created: warp_dest2src.nii.gz
Apply transfo source --> dest...
Parse list of warping fields...
Get dimensions of data...
141 x 141 x 991 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i src.nii -o src_reg.nii -t warp_src2dest.nii.gz -r dest.nii -n Linear # in /tmp/sct-20201029153546.541065-register-ocgl30mg
Copy affine matrix from destination space to make sure qform/sform are the same.
Done! To view results, type:
fsleyes dest.nii src_reg.nii &
Apply transfo dest --> source...
Parse list of warping fields...
Get dimensions of data...
512 x 448 x 26 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i dest.nii -o dest_reg.nii -t warp_dest2src.nii.gz -r src.nii -n Linear # in /tmp/sct-20201029153546.541065-register-ocgl30mg
Copy affine matrix from destination space to make sure qform/sform are the same.
Done! To view results, type:
fsleyes src.nii dest_reg.nii &
Generate output files...
File created: PAM50_t2s_reg.nii.gz
File created: 2020subj1_reg.nii
File created: warp_PAM50_t2s22020subj1.nii.gz
File created: warp_2020subj12PAM50_t2s.nii.gz
Remove temporary files...
rm -rf /tmp/sct-20201029153546.541065-register-ocgl30mg
Finished! Elapsed time: 966s
*** Generate Quality Control (QC) html report ***
Resample images to 0.6x0.6 mm
QcImage: layout with Axial slice
Compute center of mass at each slice
Mask type float64
Successfully generated the QC results in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/csvOutcome/anonData/qc/_json/qc_2020_10_29_155158.590531.json
Use the following command to see the results in a browser:
xdg-open "/media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/csvOutcome/anonData/qc/index.html"
Done! To view results, type:
fsleyes /home/nirims/sct_4.3/data/PAM50/template/PAM50_t2s.nii.gz 2020subj1_reg.nii &
Done! To view results, type:
fsleyes 2020subj1.nii PAM50_t2s_reg.nii.gz &
mv: Aufruf von stat für 'warp_t2s2PAM50_2020subj1.nii.gz' nicht möglich: Datei oder Verzeichnis nicht gefunden
--
Spinal Cord Toolbox (4.3)
Folder ../../csvOutcome/anonData/qc has been created.
Check parameters:
Working directory ........ /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
Destination image ........ 2020subj1.nii
Warping field ............ warp_template2t2s.nii.gz
Path template ............ /home/nirims/sct_4.3/data/PAM50
Output folder ............ label
WARP TEMPLATE:
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/template/PAM50_t1.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/template/PAM50_t1.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/template/PAM50_t2.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/template/PAM50_t2.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/template/PAM50_t2s.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/template/PAM50_t2s.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/template/PAM50_cord.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/template/PAM50_cord.nii.gz -n NearestNeighbor # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/template/PAM50_wm.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/template/PAM50_wm.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/template/PAM50_gm.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/template/PAM50_gm.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/template/PAM50_csf.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/template/PAM50_csf.nii.gz -n NearestNeighbor # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/template/PAM50_levels.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/template/PAM50_levels.nii.gz -n NearestNeighbor # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/template/PAM50_levels_continuous.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/template/PAM50_levels_continuous.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/template/PAM50_label_body.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/template/PAM50_label_body.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/template/PAM50_label_disc.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/template/PAM50_label_disc.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/template/PAM50_label_discPosterior.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/template/PAM50_label_discPosterior.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/template/PAM50_spine.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/template/PAM50_spine.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/template/PAM50_centerline.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/template/PAM50_centerline.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/template/PAM50_label_spinal_levels.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/template/PAM50_label_spinal_levels.nii.gz -n NearestNeighbor # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
cp /home/nirims/sct_4.3/data/PAM50/template/info_label.txt label/template
WARP ATLAS OF WHITE MATTER TRACTS:
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_00.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_00.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_01.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_01.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_02.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_02.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_03.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_03.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_04.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_04.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_05.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_05.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_06.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_06.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_07.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_07.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_08.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_08.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_09.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_09.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_10.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_10.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_11.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_11.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_12.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_12.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_13.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_13.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_14.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_14.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_15.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_15.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_16.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_16.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_17.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_17.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_18.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_18.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_19.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_19.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_20.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_20.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_21.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_21.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_22.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_22.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_23.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_23.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_24.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_24.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_25.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_25.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_26.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_26.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_27.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_27.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_28.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_28.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_29.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_29.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_30.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_30.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_31.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_31.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_32.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_32.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_33.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_33.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_34.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_34.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_35.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_35.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
/home/nirims/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /home/nirims/sct_4.3/data/PAM50/atlas/PAM50_atlas_36.nii.gz -r 2020subj1.nii -t warp_template2t2s.nii.gz -o label/atlas/PAM50_atlas_36.nii.gz -n Linear # in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/anonData/subj1
cp /home/nirims/sct_4.3/data/PAM50/atlas/info_label.txt label/atlas
*** Generate Quality Control (QC) html report ***
Resample images to 0.6x0.6 mm
QcImage: layout with Axial slice
Compute center of mass at each slice
Successfully generated the QC results in /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/csvOutcome/anonData/qc/_json/qc_2020_10_29_155605.785358.json
Use the following command to see the results in a browser:
xdg-open "/media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/csvOutcome/anonData/qc/index.html"
Done! To view results, type:
fsleyes 2020subj1.nii -cm greyscale -a 100.0 label/template/PAM50_t2.nii.gz -cm greyscale -dr 0 4000 -a 100.0 label/template/PAM50_gm.nii.gz -cm red-yellow -dr 0.4 1 -a 50.0 label/template/PAM50_wm.nii.gz -cm blue-lightblue -dr 0.4 1 -a 50.0 &
--
Spinal Cord Toolbox (4.3)
Compute shape analysis: 100%|#################| 26/26 [00:00<00:00, 47.05iter/s]
Done! To view results, type:
xdg-open /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/csvOutcome/anonData/subj1/2020subj1_gm_CSA.csv
--
Spinal Cord Toolbox (4.3)
Compute shape analysis: 100%|#################| 26/26 [00:00<00:00, 28.76iter/s]
Done! To view results, type:
xdg-open /media/sf_GemeinsamerOrdner/SCT/Testdaten/miniTrialGMWM/csvOutcome/anonData/subj1/2020subj1_wm_CSA.csv
Execution time was 1476 seconds.