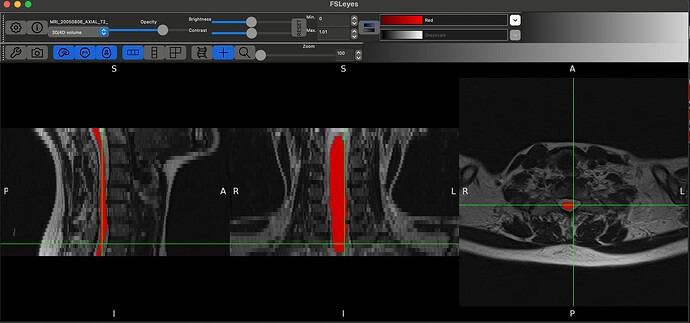
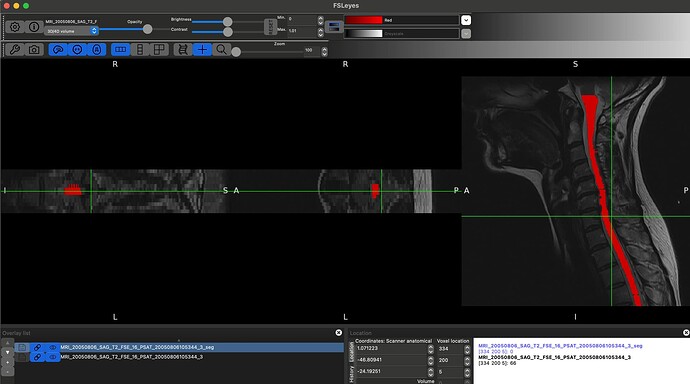
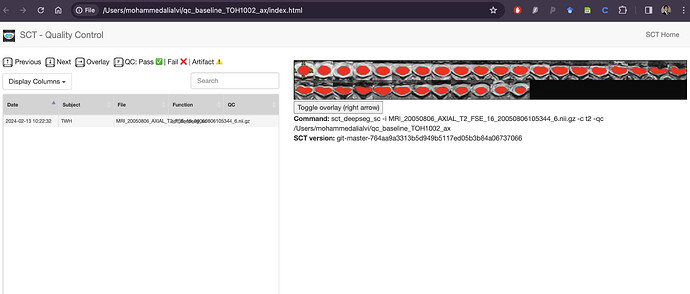
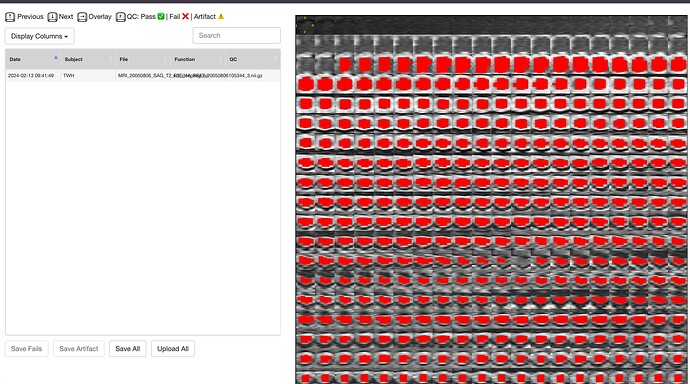
Hello @jcohenadad and @joshuacwnewton , thank you for responses above. I am a beginner myself. I have a similar question and I realize that this may be too basic. I used dcm2bids converter to convert my dicom files to nifti format with BIDS sidecars. This is what the “ls” of my target output directory for an example MRI looks like:
Directory: E:\Alvi\Thesis\tmp_dcm2bids\helper
Mode LastWriteTime Length Name
---- ------------- ------ ----
-a---- 2024-02-12 3:46 PM 911 002_TOH1002_COR_LOC.json
-a---- 2024-02-12 3:46 PM 1493545 002_TOH1002_COR_LOC.nii.gz
-a---- 2024-02-12 3:46 PM 1001 003_TOH1002_SAG_T2_FSE_16_PSAT.json
-a---- 2024-02-12 3:46 PM 3736351 003_TOH1002_SAG_T2_FSE_16_PSAT.nii.gz
-a---- 2024-02-12 3:46 PM 973 004_TOH1002_SAG_T1_FSE_2_PSAT.json
-a---- 2024-02-12 3:46 PM 3784015 004_TOH1002_SAG_T1_FSE_2_PSAT.nii.gz
-a---- 2024-02-12 3:46 PM 995 005_TOH1002_AXIAL_T2_FST_VOL_MTC_PSAT.json
-a---- 2024-02-12 3:46 PM 6321888 005_TOH1002_AXIAL_T2_FST_VOL_MTC_PSAT.nii.gz
-a---- 2024-02-12 3:46 PM 939 006_TOH1002_AXIAL_T2_FSE_16.json
-a---- 2024-02-12 3:46 PM 1893630 006_TOH1002_AXIAL_T2_FSE_16.nii.gz
As you can, see converting the whole DICOM folder to NIFTI generates these individual sequences. How can I use these files to then input into sct_deepseg_sc?
Again, I apologize if this is too basic or if this is not the right place to ask this?