dear jcohenadad
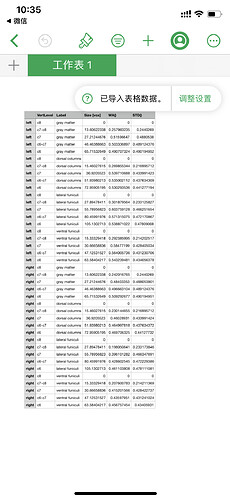
these are list of labels
0, WM left fasciculus gracilis, PAM50_atlas_00.nii.gz
1, WM right fasciculus gracilis, PAM50_atlas_01.nii.gz
2, WM left fasciculus cuneatus, PAM50_atlas_02.nii.gz
3, WM right fasciculus cuneatus, PAM50_atlas_03.nii.gz
4, WM left lateral corticospinal tract, PAM50_atlas_04.nii.gz
5, WM right lateral corticospinal tract, PAM50_atlas_05.nii.gz
6, WM left ventral spinocerebellar tract, PAM50_atlas_06.nii.gz
7, WM right ventral spinocerebellar tract, PAM50_atlas_07.nii.gz
8, WM left rubrospinal tract, PAM50_atlas_08.nii.gz
9, WM right rubrospinal tract, PAM50_atlas_09.nii.gz
10, WM left lateral reticulospinal tract, PAM50_atlas_10.nii.gz
11, WM right lateral reticulospinal tract, PAM50_atlas_11.nii.gz
12, WM left spinal lemniscus (spinothalamic and spinoreticular tracts), PAM50_atlas_12.nii.gz
13, WM right spinal lemniscus (spinothalamic and spinoreticular tracts), PAM50_atlas_13.nii.gz
14, WM left spino-olivary tract, PAM50_atlas_14.nii.gz
15, WM right spino-olivary tract, PAM50_atlas_15.nii.gz
16, WM left ventrolateral reticulospinal tract, PAM50_atlas_16.nii.gz
17, WM right ventrolateral reticulospinal tract, PAM50_atlas_17.nii.gz
18, WM left lateral vestibulospinal tract, PAM50_atlas_18.nii.gz
19, WM right lateral vestibulospinal tract, PAM50_atlas_19.nii.gz
20, WM left ventral reticulospinal tract, PAM50_atlas_20.nii.gz
21, WM right ventral reticulospinal tract, PAM50_atlas_21.nii.gz
22, WM left ventral corticospinal tract, PAM50_atlas_22.nii.gz
23, WM right ventral corticospinal tract, PAM50_atlas_23.nii.gz
24, WM left tectospinal tract, PAM50_atlas_24.nii.gz
25, WM right tectospinal tract, PAM50_atlas_25.nii.gz
26, WM left medial reticulospinal tract, PAM50_atlas_26.nii.gz
27, WM right medial reticulospinal tract, PAM50_atlas_27.nii.gz
28, WM left medial longitudinal fasciculus, PAM50_atlas_28.nii.gz
29, WM right medial longitudinal fasciculus, PAM50_atlas_29.nii.gz
30, GM left ventral horn, PAM50_atlas_30.nii.gz
31, GM right ventral horn, PAM50_atlas_31.nii.gz
32, GM left intermediate zone, PAM50_atlas_32.nii.gz
33, GM right intermediate zone, PAM50_atlas_33.nii.gz
34, GM left dorsal horn, PAM50_atlas_34.nii.gz
35, GM right dorsal horn, PAM50_atlas_35.nii.gz
36, CSF contour, PAM50_atlas_36.nii.gz
50, spinal cord, 0:35
51, white matter, 0:29
52, gray matter, 30:35
53, dorsal columns, 0:3
54, lateral funiculi, 4:13
55, ventral funiculi, 14:29
56, left hemi-cord, 0,2,4,6,8,10,12,14,16,18,20,22,24,26,28,30,32,34
57, right hemi-cord, 1,3,5,7,9,11,13,15,17,19,21,23,25,27,29,31,33,35
there is no label for gm left lateral and gm right lateral . if there are other labels ?