Hi @Caroline_Landelle
Thank you for reaching out.
1/ I wonder if there is a solution to avoid two steps to keep the resolution of the func image after the coregistration (sct_register_multimodal + sct_resample).
I’m not sure I fully understand what you are trying to achieve. Firstly, there is no need to resample the source (ie: ‘moving’) image if it will ultimately be resampled to the destination space (ie: ‘fixed’ image), which IIUC is the high-resolution anatomical image. So indeed, you could probably skip the sct_resample stage. Secondly, sct_register_multimodal already includes a resampling stage: after estimating the forward and backward warping fields, the source image is registered and consequently resampled to the destination image (with suffix ‘_reg’).
Is there a possibility with sct_register_multimodal to say “do not change the resolution of the input image”?
This function outputs the forward (source → dest) and backward (dest → source) warping fields, so if you wish to concatenate them with your pipeline, then you can do so and not use the source_reg image at this stage. But again, I am unsure what you are trying to achieve so it is difficult for me to advise on best practice. All I can say, is that in general fMRI data should be analyzed (first level analysis) in their native space so as to minimize interpolation errors, and what is warped to the anatomical image and/or template is the results of the first-level analysis (eg z-score maps).
2/ I can’t quite explain that my tSNR (func) in axial plane increases after coregistration especially in the GM (which is good). Do you think it’s an interpolation effect related to the coregistration step or something like that?
Interpolating (with trilinear, sinc methods) a time series changes noise properties so it is not surprising that the tSNR goes up. TSNR should always be computed on the native image space. If you with to perform motion correction before computing tSNR, then I suggest you use the nearest neighbor interpolation (flag: -x nn).
3/ Finally, do you think it is possible to get a deformation field file after moco? The idea is to apply a single transformation (sct_apply_transfo) to all my func volumes by concatenating (sct_concat_transfo) all the warping fields (e.g. I will concatenate warp_func_to_func_moco + warp_fun_moco_to_anat + warp_anat_to_PAM50 = warp_func_to_PAM50).
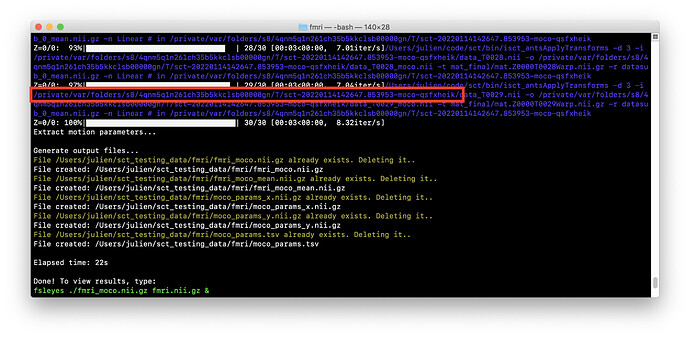
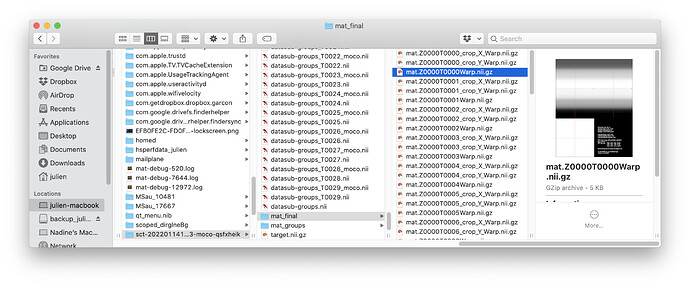
You can access these by setting the flag -r 0 in sct_fmri_moco. You can locate the temporary folder in your terminal, example:
And then you can access the intermediate warping fields for each volume of the 4D time series:
This post could also be helpful to you.
However, as said earlier, in general fMRI scans should be analyzed in their native space to minimize interpolation errors (eg: modification of tSNR you observed at point #2) and keeping the proper degree of freedom when using spatial autocorrelation regularization methods for adjusting p-values using Gaussian random field approaches.