Hello,
I’ve been trying to apply some sct methods on ex vivo spinal cord data of rats.
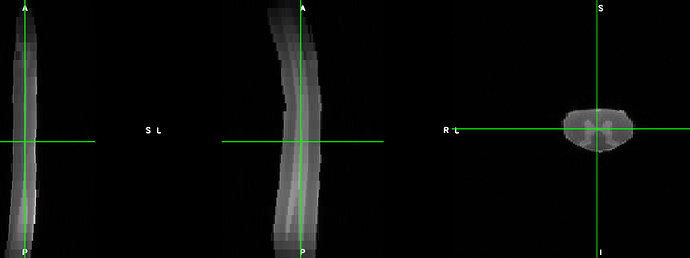
The t2 images look like this:
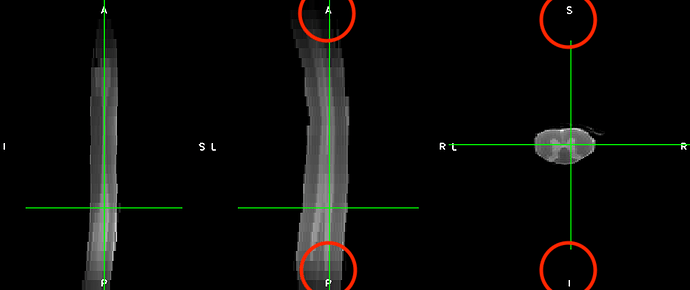
I managed to run sct_propseg on it to obtain the centerline and segmentation mask. This worked quite well.
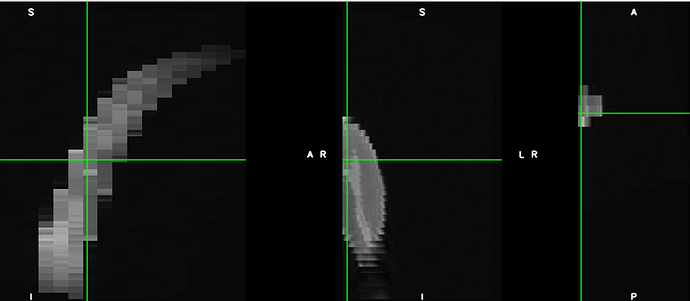
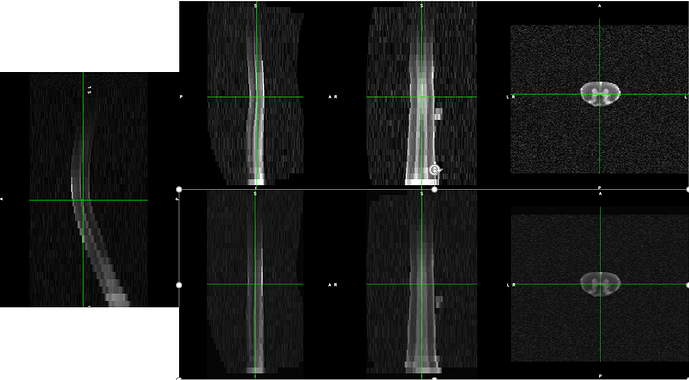
However, when I try to straigthen them with sct_straighten_spinalcord, I keep getting something like this:
I attached a zip file with the example above, and the output from sct_propseg.
You can reproduce the problem with the command
sct_straighten_spinalcord -i vol0000.nii.gz -s vol0000_seg.nii.gz
I tried using the centerline or the segmentation as input argument, I tried hanning/nurbs algorithm, and played around with some bspline parameters, none of the parameters seems to improved the result.
Could there be some fundamental problem with my dataset (unusual dimensions or orientation)?
One important note: sct_propseg only worked when I used the argument “-rescale 3” which was mentioned in the mouse pipeline. Unfortunately, sct_straighten_spinalcord doesn’t have such a parameter.
Thank you very much
rat_example.7z (1.2 MB)