Dear Forum
I have, for some time, been playing around with some data, trying to find out whether frequency navigators (in Siemens parlance, “Phase Stabilisation”) are worth employing in cervical spinal cord imaging using gradient-echo sequences. To the best of my knowledge (and please correct me if I am wrong, I am by no means the expert), such an evaluation has not been done before.
With @jcohenadad 's post-hoc permission, I have linked this thread in my OHBM poster on this topic. Easier to discuss things on here.
In short, I think it is worth turning this feature on (if it is provided by your vendor).
However, since this is only a poster, not a full paper, I am not sure that we did the best investigation.
The results I am basing my recommendation on are a consistent trend, but at least in the way we looked at them, not significant. Caveat lector.
To go a bit more in depth, read further.
1, Why is some correction needed?
The spine is surrounded by many tissue and air/tissue boundaries, vessels, etc, etc. In MR physics terms, it is surrounded by sources of phase that are not internal to the spinal cord itself. In turn, this additional phase term will manifest in the magnitude images used in MR imaging as a loss of contrast and a generally fuzzier, blurrier picture.
This will make image operations, such as segmentation, harder, which then might influence the research or clinical outcome. Here, I will make no claim to the exact nature of the phase difference, nor of its exact source (flow, physiological motion, susceptibility boundaries, etc), just present the problem and a way to deal with it.
2, What can be done to correct for this?
Thankfully, this is an easy problem to solve. I apologise for the techspeak you are about to read.
Within a TR, the phase we measure can be split into the phase coming from the tissue in question (eg, the phase measured in a voxel is caused by things in that voxel), and the outside sources of phase (eg, the phase measured in a voxel is coming from things outside that voxel).
The phase coming form within the voxel should be constant over time (at least within the cord), while the outside sources are not.
Thus, if we measure the same voxel over and over again (in practical terms, acquire the central k-space line over and over again), then we can separate the phase that comes from inside the voxel/line of voxels from that coming from other sources. With the reasonable assumption that we start from a position where the outside sources have not had an effect (because we did not give them time to build up and influence our signal), we can measure the local tissue contribution in the first TR, and then every time we measure the central line again, we measure both the local and nonlocal effect, and can then subtract the nonlocal effect from other measurements in the same TR.
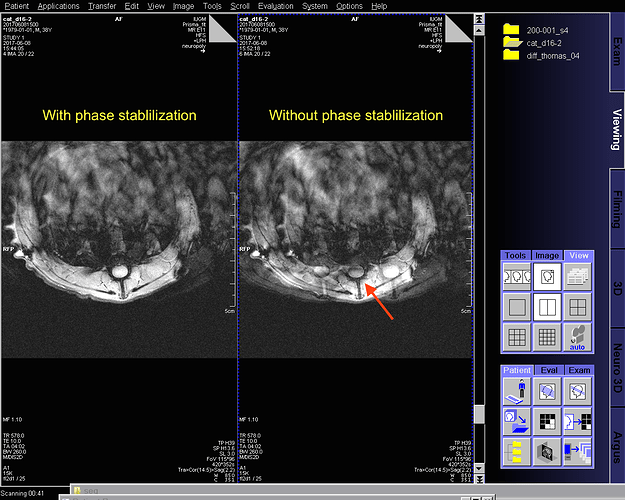
If you have a Siemens scanner, this approach is called “Phase Stabilisation”. In scanners running VD or earlier, this checkbox is visible to the user when running a GRE sequence. However, on VE, this option was removed from the GUI, while the underlining code has been kept in place. If you have access to an IDEA platform, you can look in detail at the process in your Ice Users Guide document.
Crucially, I have no information on why this was removed, so there may be a good reason for it.
3, Our outcome metric
Again, I am not sure that this is the best way to measure this. However, the following idea seemed to be closest to something clinically relevant.
We assume that the issue detailed in the first point results in a lack of contrast and less well defined borders between the spinal cord and the CSF, and between grey and white matter. The downstream effect of this is that the segmentation of CSF/SC and GM/WM should be in some way different between scans acquired with or without this navigator. Comparing to a gold standard, I would expect the segmentation of the scans with the navigator to be better than those without. A simple way to check this was to look at the cross-sectional area (CSA) of the spinal cord and the grey matter.
4, Methods: Acquisition
This is a little side project that popped up as part of a bigger study (currently being written up), so I did not tailor my scan parameters for this specifically.
We acquired three scans of the cervical spinal cord (minimum coverage of C2-C5), positioned the FOV according to Julien’s SOP, used the same shim volume, and so forth.
The three scans were a MEDIC scan as gold standard and two multi-echo GRE scans with MT contrast, one with and one without the navigator technique. The two MT scans (MT_Nav and MT_NoNav) had matched TR, TEs, echoes, echo spacing, and only differed in wether or not the navigator was acquired. In-plane resolution for the MT scans was 625um^2, and 500um^2 for the MEDIC. Effective TE for the MEDIC was 20ms, and the last TE of the MT scans was ~17ms.
We did not save the individual echoes of the MEDIC scan, nor did we (or could we) turn on the navigator functionality for it.
We imaged 10 subjects, all healthy compliant volunteers between 24 and 44 (the usual “local postdocs and colleagues” cohort).
5, Methods: Processing
The two MT scans were co-registered to the MEDIC scan using SCT (4.2), using the rigid body algorithm. Thus, the anatomy should be matched, but without any nonlinear deformations that may counteract the effect we wanted to investigate. While this step did include an upsampling, and thus may have introduced a bias, that bias should at least be consistent between the two MT scans.
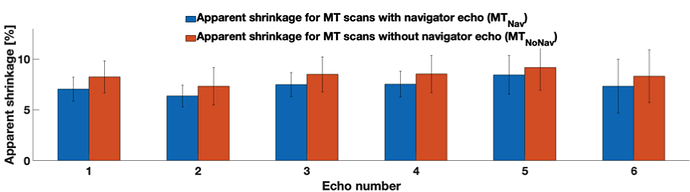
Next, we segmented the spinal cord and the grey matter from all three of these scans (again SCT 4.2, deepseg algorithm for both), and computed what I call “apparent shrinkage”, which is the normalised difference between the CSA of the MT scan and the MEDIC scan, summed over all but the outermost slices of the acquisition:
((CSA_MT-CSA_MEDIC)/CSA_MEDIC)*100%
Eg it answers this question: How much smaller does the segmented spinal cord/grey matter appear to be due to the lack of frequency navigator?
6, Results:
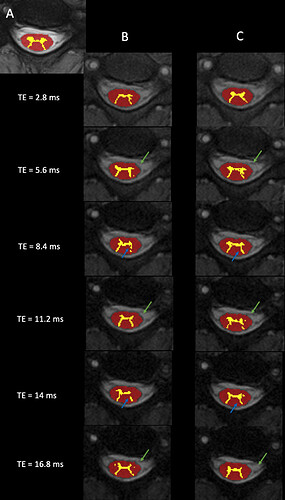
Just looking at it visually, both cord and grey matter segmentation is improved using this approach. A is the MEDIC, B are the MTw scans without the navigator, C are with.
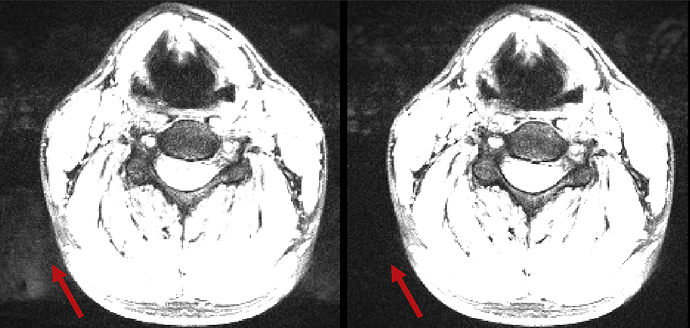
Additionally, the characteristic GRAPPA artefacts are reduced both inside and outside the neck, because the ususal phase mismatch between the GRAPPA kernel and the rest of the acquisition is mitigated.
The apparent shrinkage is reduced by the navigator, but the difference is sadly not significant:
7, What could/should have been done differently
As I said, this was a little side-project. If I were to run this again, my two main changes would be:
A, Only acquire one MTw scan, one with a navigator, and reconstruct the data offline, both using and not using the navigator correction. This way, I would not have to contend with the problem of motion between scans. While I am confident that the effect I show is not just down to the volunteers moving, and thus the slices being slightly different, I can not be 100% sure.
B, We did not have access to the MEDIC scan code, nor have we saved the raw data. It would be interesting to see if the MEDIC, used/recommended as a gold standard in many places, can be improved by tagging this on.
I’m happy to forge future collaborations, if you are interested in exploring this further, as I have a few other ideas.
I’m happy to answer any questions and grateful for any feedback!
Edit: I’d like to thank Johanna Vannesjö and Julien for the off-line discussions that prompted me to look into this. Apologies for not including you in the original post.