Hello,
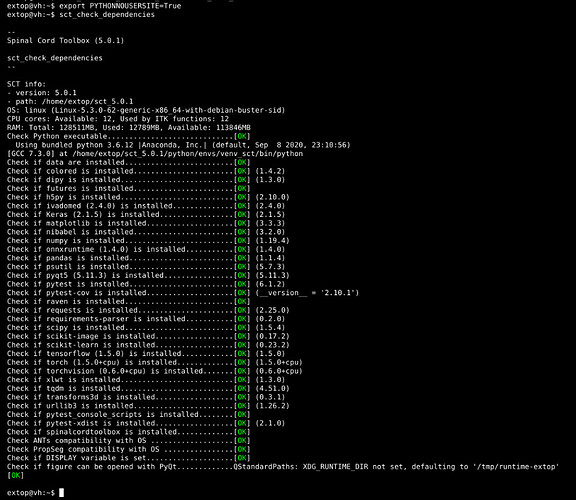
Here the sct_check_dependencies that went weird
extop@:/mnt/aran/testing/001_1$ sct_check_dependencies
--
Spinal Cord Toolbox (5.0.0)
SCT info:
- version: 5.0.0
- path: /home/extop/sct_5.0.0
OS: linux (Linux-5.3.0-62-generic-x86_64-with-debian-buster-sid)
CPU cores: Available: 12, Used by ITK functions: 12
RAM: Total: 128511MB, Used: 20537MB, Available: 106404MB
Check Python executable.............................[OK]
Using bundled python 3.6.12 |Anaconda, Inc.| (default, Sep 8 2020, 23:10:56)
[GCC 7.3.0] at /home/extop/sct_5.0.0/python/envs/venv_sct/bin/python
Check if pytest-cov is installed....................[OK] (__version__ = '2.10.1')
Check if requests is installed......................[OK] (2.24.0)
Check if requirements-parser is installed...........[OK] (0.2.0)
Check if scipy is installed.........................[OK] (1.5.4)
Check if scikit-image is installed..................[OK] (0.17.2)
Check if scikit-learn is installed..................[OK] (0.23.2)
Check if tensorflow (1.5.0) is installed............[OK] (1.5.0)
Check if torch (1.5.0+cpu) is installed.............[WARNING] (1.6.0 != 1.5.0+cpu mandated version))
Check if torchvision (0.6.0+cpu) is installed.......[WARNING] (0.7.0 != 0.6.0+cpu mandated version))
Check if xlwt is installed..........................[OK] (1.3.0)
Check if tqdm is installed..........................[OK] (4.51.0)
Check if transforms3d is installed..................[OK] (0.3.1)
Check if urllib3 is installed.......................[OK] (1.25.11)
Check if pytest_console_scripts is installed........[OK]
Check if pytest-xdist is installed..................[OK] (2.1.0)
Check if figure can be opened with PyQt.............[OK]
And then, the sct_testing
extop@:/mnt/aran/testing/001_1$ sct_testing
--
Spinal Cord Toolbox (5.0.0)
Trying URL: https://github.com/sct-data/sct_testing_data/releases/download/r20201030/sct_testing_data-r20201030.zip
Downloading: sct_testing_data-r20201030.zip
Status: 100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 7.17M/7.17M [00:12<00:00, 595kB/s]
Creating temporary folder (/tmp/sct-20201113075848.995069-2wp8i281)
Unzip data to: /tmp/sct-20201113075848.995069-2wp8i281
Removing temporary folders...
Done!
Creating temporary folder (/tmp/sct-20201113075849.064687-gpcy0fls)
Will run through the following tests:
- sequentially: sct_deepseg_gm sct_deepseg_lesion sct_deepseg_sc
- in parallel with 12 jobs: sct_analyze_lesion sct_analyze_texture sct_apply_transfo sct_convert sct_compute_ernst_angle sct_compute_hausdorff_distance sct_compute_mtr sct_compute_mscc sct_compute_snr sct_create_mask sct_crop_image sct_dice_coefficient sct_detect_pmj sct_dmri_compute_dti sct_dmri_concat_b0_and_dwi sct_dmri_concat_bvals sct_dmri_concat_bvecs sct_dmri_compute_bvalue sct_dmri_moco sct_dmri_separate_b0_and_dwi sct_dmri_transpose_bvecs sct_extract_metric sct_flatten_sagittal sct_fmri_compute_tsnr sct_fmri_moco sct_get_centerline sct_image sct_label_utils sct_label_vertebrae sct_maths sct_merge_images sct_process_segmentation sct_propseg sct_qc sct_register_multimodal sct_register_to_template sct_resample sct_smooth_spinalcord sct_straighten_spinalcord sct_warp_template
Checking sct_deepseg_gm.............................[FAIL]
====================================================================================================
sct_deepseg_gm -i t2s/t2s_uncropped.nii.gz -o output.nii.gz -qc testing-qc
====================================================================================================
--
Spinal Cord Toolbox (5.0.0)
Traceback (most recent call last):
File "/home/extop/sct_5.0.0/scripts/sct_deepseg_gm.py", line 138, in <module>
run_main()
File "/home/extop/sct_5.0.0/scripts/sct_deepseg_gm.py", line 121, in run_main
use_tta)
File "/home/extop/sct_5.0.0/spinalcordtoolbox/deepseg_gm/deepseg_gm.py", line 313, in segment_file
use_tta)
File "/home/extop/sct_5.0.0/spinalcordtoolbox/deepseg_gm/deepseg_gm.py", line 233, in segment_volume
deepgmseg_model.load_weights(model_abs_path)
File "/home/extop/sct_5.0.0/python/envs/venv_sct/lib/python3.6/site-packages/keras/engine/topology.py", line 2656, in load_weights
f, self.layers, reshape=reshape)
File "/home/extop/sct_5.0.0/python/envs/venv_sct/lib/python3.6/site-packages/keras/engine/topology.py", line 3328, in load_weights_from_hdf5_group
original_keras_version = f.attrs['keras_version'].decode('utf8')
AttributeError: 'str' object has no attribute 'decode'
Checking sct_deepseg_lesion.........................[FAIL]
====================================================================================================
sct_deepseg_lesion -i t2/t2.nii.gz -c t2
====================================================================================================
--
Spinal Cord Toolbox (5.0.0)
Method:
Centerline algorithm: svm
Assumes brain section included in the image: True
Creating temporary folder (/tmp/sct-20201113075855.108779-xyc7gjs3)
Reorient the image to RPI, if necessary...
Finding the spinal cord centerline...
Creating temporary folder (/tmp/sct-20201113075855.446297-yc033ooj)
Remove temporary files...
rm -rf /tmp/sct-20201113075855.446297-yc033ooj
Cropping the image around the spinal cord...
Normalizing the intensity...
load data...
Segmenting the MS lesions using deep learning on 3D patches...
Traceback (most recent call last):
File "/home/extop/sct_5.0.0/scripts/sct_deepseg_lesion.py", line 176, in <module>
main()
File "/home/extop/sct_5.0.0/scripts/sct_deepseg_lesion.py", line 152, in main
brain_bool=brain_bool, remove_temp_files=remove_temp_files, verbose=verbose)
File "/home/extop/sct_5.0.0/spinalcordtoolbox/deepseg_lesion/core.py", line 201, in deep_segmentation_MSlesion
im=im_res3d.copy())
File "/home/extop/sct_5.0.0/spinalcordtoolbox/deepseg_lesion/core.py", line 88, in segment_3d
seg_model = load_trained_model(model_fname)
File "/home/extop/sct_5.0.0/spinalcordtoolbox/deepseg_sc/cnn_models_3d.py", line 91, in load_trained_model
return load_model(model_file, custom_objects=custom_objects)
File "/home/extop/sct_5.0.0/python/envs/venv_sct/lib/python3.6/site-packages/keras/models.py", line 242, in load_model
model_config = json.loads(model_config.decode('utf-8'))
AttributeError: 'str' object has no attribute 'decode'
Checking sct_deepseg_sc.............................[FAIL]
====================================================================================================
sct_deepseg_sc -i t2/t2.nii.gz -c t2 -qc testing-qc
====================================================================================================
--
Spinal Cord Toolbox (5.0.0)
Config deepseg_sc:
Centerline algorithm: svm
Brain in image: True
Kernel dimension: 2d
Contrast: t2
Threshold: 0.7
Creating temporary folder (/tmp/sct-20201113075858.725650-0mpax34n)
Reorient the image to RPI, if necessary...
Finding the spinal cord centerline...
Creating temporary folder (/tmp/sct-20201113075858.980012-fde3c2iu)
Remove temporary files...
rm -rf /tmp/sct-20201113075858.980012-fde3c2iu
Cropping the image around the spinal cord...
Normalizing the intensity...
Segmenting the spinal cord using deep learning on 2D patches...
Traceback (most recent call last):
File "/home/extop/sct_5.0.0/scripts/sct_deepseg_sc.py", line 208, in <module>
main()
File "/home/extop/sct_5.0.0/scripts/sct_deepseg_sc.py", line 192, in main
threshold_seg=threshold, remove_temp_files=remove_temp_files, verbose=verbose)
File "/home/extop/sct_5.0.0/spinalcordtoolbox/deepseg_sc/core.py", line 509, in deep_segmentation_spinalcord
im_in=im_norm_in)
File "/home/extop/sct_5.0.0/spinalcordtoolbox/deepseg_sc/core.py", line 350, in segment_2d
seg_model.load_weights(model_fname)
File "/home/extop/sct_5.0.0/python/envs/venv_sct/lib/python3.6/site-packages/keras/engine/topology.py", line 2656, in load_weights
f, self.layers, reshape=reshape)
File "/home/extop/sct_5.0.0/python/envs/venv_sct/lib/python3.6/site-packages/keras/engine/topology.py", line 3328, in load_weights_from_hdf5_group
original_keras_version = f.attrs['keras_version'].decode('utf8')
AttributeError: 'str' object has no attribute 'decode'
Checking sct_analyze_lesion.........................[OK]
Checking sct_analyze_texture........................[OK]
Checking sct_apply_transfo..........................[OK]
Checking sct_convert................................[OK]
Checking sct_compute_ernst_angle....................[OK]
Checking sct_compute_hausdorff_distance.............[OK]
Checking sct_compute_mtr............................[OK]
Checking sct_compute_mscc...........................[OK]
Checking sct_compute_snr............................[OK]
Checking sct_create_mask............................[OK]
Checking sct_crop_image.............................[OK]
Checking sct_dice_coefficient.......................[OK]
Checking sct_detect_pmj.............................[OK]
Checking sct_dmri_compute_dti.......................[OK]
Checking sct_dmri_concat_b0_and_dwi.................[OK]
Checking sct_dmri_concat_bvals......................[OK]
Checking sct_dmri_concat_bvecs......................[OK]
Checking sct_dmri_compute_bvalue....................[OK]
Checking sct_dmri_moco..............................[OK]
Checking sct_dmri_separate_b0_and_dwi...............[OK]
Checking sct_dmri_transpose_bvecs...................[OK]
Checking sct_extract_metric.........................[OK]
Checking sct_flatten_sagittal.......................[OK]
Checking sct_fmri_compute_tsnr......................[OK]
Checking sct_fmri_moco..............................[OK]
Checking sct_get_centerline.........................[OK]
Checking sct_image..................................[OK]
Checking sct_label_utils............................[OK]
Checking sct_label_vertebrae........................[OK]
Checking sct_maths..................................[OK]
Checking sct_merge_images...........................[OK]
Checking sct_process_segmentation...................[OK]
Checking sct_propseg................................[OK]
Checking sct_qc.....................................[OK]
Checking sct_register_multimodal....................[OK]
Checking sct_register_to_template...................[OK]
Checking sct_resample...............................[OK]
Checking sct_smooth_spinalcord......................[OK]
Checking sct_straighten_spinalcord..................[OK]
Checking sct_warp_template..........................[OK]
status: [1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]
Failures: sct_deepseg_gm sct_deepseg_lesion sct_deepseg_sc
Finished! Elapsed time: 129s