Continuing the discussion from Account temporarily on hold:
Dear SCT team:
I am currently working on the processing of spinal cord DTI data. Before the processing of spinal cord data, I first preprocessed the DTI data (including Topup, eddycorrect, DTIFIT), and then processed the spinal cord data according to the tutorial in Beijing in August 2019. The steps are as follows:
1:spinalcord segment;
2:create a mask;
3:crop image;
4:motion correction;
5:After doing the four steps above I perform the final segmentation of the spinal cord:
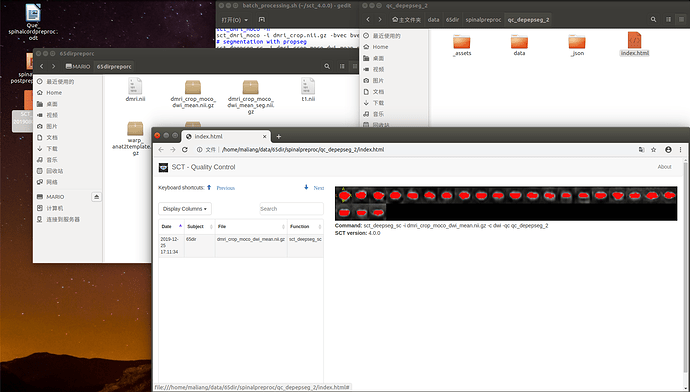
command: sct_deepseg_sc -i dmri_crop_moco_dwi_mean.nii.gz -c dwi -qc qc_deepseg ~~~
but my segment result was not very good,this is my first question and i will attach it in attachments.
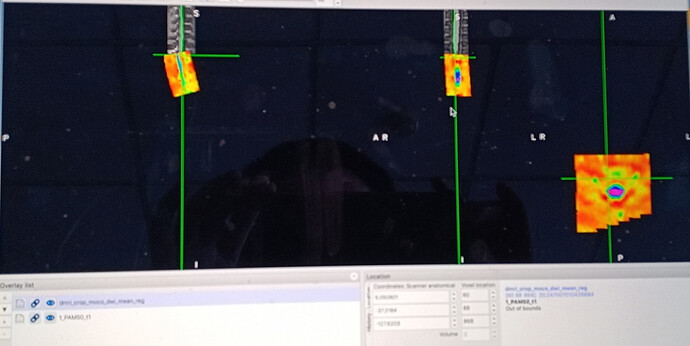
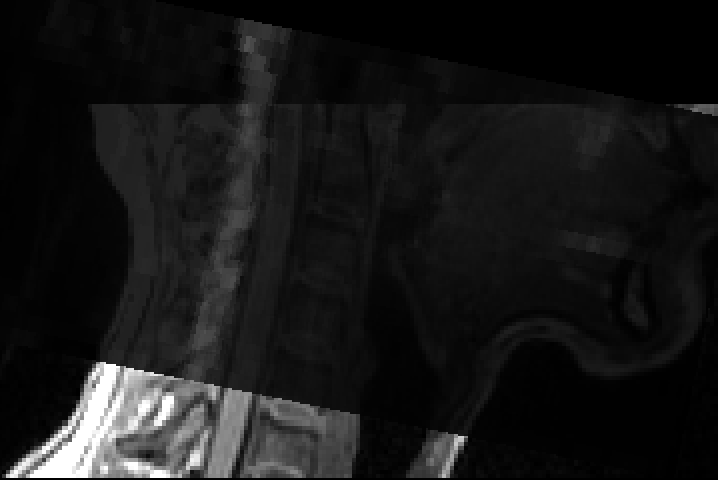
the second question is when I conducted spinal cord registration, and the order of spinal cord registration was as follows:
Sct_register_multimodal -i $SCT_DIR/data/PAM50 / template/PAM50_t1 nii. Gz iseg - $SCT_DIR/data/PAM50 / template/PAM50_cord nii. Gz - d dmri_crop_moco_dwi_mean. Nii. Gz - dseg dmri_crop_moco_dwi_mean_seg. nii. gz- param step = 1, type = seg, algo = centermass: step = 2, type = seg, algo = bsplinesyn, slicewise = 1, 3 - initwarp iter =…/ t1 / warp_template2anat. Nii. Gz - initwarpinv…/ t1 / warp_anat2template. nii. gz - qc qc_reg
The result of registration is completely distorted, I suspect the reason is warp, could you please help me solve this problem,I will put the required documents in the attachment.A thousand thanks.
my version is SCT_v4.0.0 in ubuntu 16.04,because the attachment is too big ,may i have your email and email it to you for your convenience to find the problem
Sincerely!