Hi @jcohenadad!
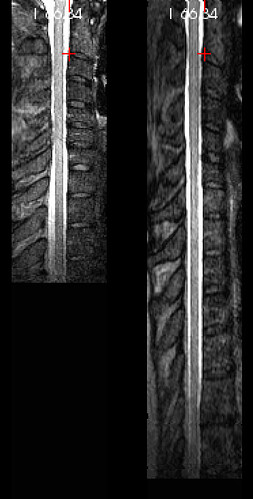
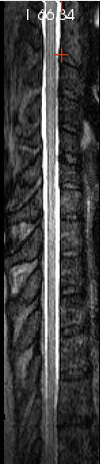
I re-ran my preprocessing pipeline on a dataset that I previously processed a couple months ago, and noticed that the result was odd. (For cervical regions, my pipeline has the user define only label “3”.) Upon closer inspection (pic below), it was clear that the vertebral levels were stretched about 50% further than they should have been (shown on the right vs. a proper warping field from another subject on the left).
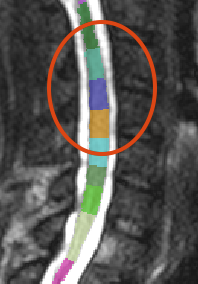
I am, of course, aware of this very helpful video showing the selection of multiple cervical vertebral levels:
… as well as this topic:
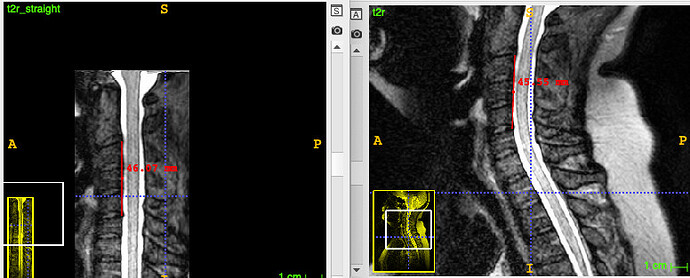
However, I had an issue just over a year ago with the warping of some thoracic data, and the solution was to define only two thoracic points instead of as many as possible (albeit with my error of overlabeling past “21”):
Therefore, I have to admit that I am uncertain as to what the best practices are for manually defining vertebral levels (hence the title of this post ![]() ). There are examples of labeling all cervical levels (which I presume would fix the incorrect cervical warping field that I just observed), but there is also the issue with thoracolumbar labeling where the warping field was less “funky” when only two points were labeled (which, to be honest, I still don’t quite understand).
). There are examples of labeling all cervical levels (which I presume would fix the incorrect cervical warping field that I just observed), but there is also the issue with thoracolumbar labeling where the warping field was less “funky” when only two points were labeled (which, to be honest, I still don’t quite understand).
I am processing dozens of cervical or thoracolumbar datasets and my #1 wish, of course, is that the results are as accurate as possible (and stable across SCT versions). I have absolutely no problem manually defining each level for the cervical and thoracic scans if that is the best path forward for the results to be as accurate as possible; however, I didn’t write this code and don’t want to make assumptions (e.g., the issue of the thoracic warping field discussed previously). Would you be able to comment on the best practices for manually defining cervicothoracolumbar vertebral levels? Thanks! ![]()
-Rob