Hi, I am in the middle of preprocessing of an EPI image of the spinal cord and I am facing a bug that prevents sct_fmri_moco from working properly, and I am posting this to ask for help in dealing with the problem. As you can see in the attached video, the EPI image after sct_fmri_moco is done is moving a lot per volume. the same EPI image just before sct_fmri_moco does not show such a big movement.
I currently run my scrip in the image preprocessing, and I do not see such abnormalities after sct_fmri_moco in the EPI images of other runs of the same subject, so I think it is probably not a mistake of my script.
Have you ever had any experience with such a bug? If so, please let me know how to deal with it.
The command I am using is listed below.
sct_fmri_moco -i “${input_file}.nii.gz” -m ${Mask} -x spline
I’m looking forward to your reply.
Hi,
This issue is likely caused by an important motion occurring during the scan, which asks for some modification of the default parameters. Notably:
- increase mask size
- changing grouping option
- changing degree of polynomial function
- changing metric for the cost function
I can play around with these parameters if you send me (privately) the dataset with your current mask.
Best,
Julien
Hi, Dr. Cohen-Adad.
Thank you for your reply.
I have sent the relevant files to your address listed in the SCT.
I hope you resolve this issue.
Hi,
You did not send me your mask file (only the fMRI time series with/without motion correction). So I wrote some code to get the mask, followed by a syntax for motion correction (moco) that seems to do a decent job for these data. Note that I also suggest to crop the fMRI data for faster processing (without cropping: 1338s, with cropping: 96s).
# Average time series across time (to perform cord segmentation)
sct_maths -i fcn_rmvol_mcf_ref_SP.nii.gz -mean t -o fcn_rmvol_mcf_ref_SP_mean.nii.gz
# Coarse segmentation of the spinal cord (to create a mask for moco)
sct_deepseg_sc -i fcn_rmvol_mcf_ref_SP_mean.nii.gz -c dwi
# Create a mask around the spinal cord (to improve moco performance)
sct_create_mask -i fcn_rmvol_mcf_ref_SP_mean.nii.gz -p centerline,fcn_rmvol_mcf_ref_SP_mean_seg.nii.gz -f cylinder -size 35mm
# Crop EPI time series for faster processing
sct_crop_image -i fcn_rmvol_mcf_ref_SP.nii.gz -m mask_fcn_rmvol_mcf_ref_SP_mean.nii.gz -o fcn_rmvol_mcf_ref_SP_crop.nii.gz
# Also crop the segmentation so we can use it for the quality control (QC)
sct_crop_image -i fcn_rmvol_mcf_ref_SP_mean_seg.nii.gz -m mask_fcn_rmvol_mcf_ref_SP_mean.nii.gz -o fcn_rmvol_mcf_ref_SP_mean_seg_crop.nii.gz
# Motion correction
sct_fmri_moco -i fcn_rmvol_mcf_ref_SP_crop.nii.gz -m mask_fcn_rmvol_mcf_ref_SP_mean.nii.gz -ofolder mask_crop -qc qc -qc-seg fcn_rmvol_mcf_ref_SP_mean_seg_crop.nii.gz
Here is the quality control report (unzip, and open the index.html file in your browser).
You notice that adjacent slices tend to slightly move in opposite direction: this is likely caused by the slice acquisition mode that was set to ‘interleaved’. In this case, due to the breathing that creates a slight offset, the odd slices are not experiencing the same B0 field as the even slices, creating this ‘zigzag’ pattern along the superior inferior direction. I usually recommend acquiring axial time series (fMRI or DWI) with ‘ascending’ or ‘descending’ mode, so that the breathing-induced B0 offset appears gradually along the S-I direction, and is more easily accounted for by the motion correction algorithm (because the latter is highly regularized along the S-I direction).
Hope that helps,
Julien
Hi, thanks for the advice.
I looked over my email and I did send the mask image. Did you not receive it?
So you think that this sct_fmri_moco bug is caused by the fact that the slice acquisition mode is set to “interleaved” at the stage of acquiring the EPI image, and that it is difficult to correct this in preprocessing?
Then, let’s change the slice acquisition mode to “ascending” or “descending” as a trial. Thank you very much.
Indeed, you did send the mask and I missed it, my apologies.
When using your mask, I cannot reproduce the issue you showed in your video. Here is the result of motion correction using your mask:
And here is the syntax I used:
sct_fmri_moco -i fcn_rmvol_mcf_ref_SP.nii.gz -m mask_ref_SP_23.nii.gz
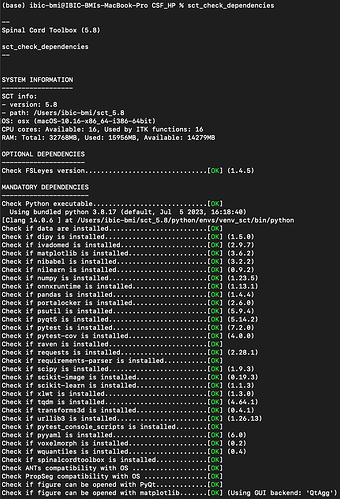
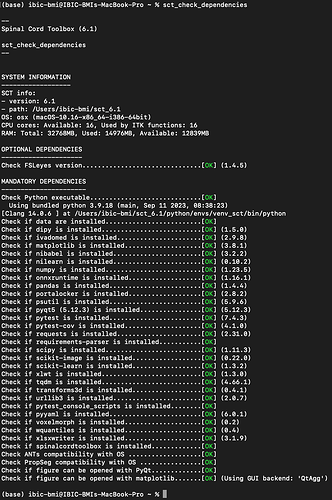
So maybe you are using a different SCT version than me (I am on the latest). Can you please run the command below and copy/paste the output:
sct_check_dependencies
Another thing I noticed, is that the data you showed in your video does not look like the data you sent me. In order for me to reproduce your issue, I need the exact same data, because the issue might be data-specific.
No, this is not what I meant. I don’t know what causes the issue you are experiencing because I cannot reproduce it (let’s see what version of SCT you are using, to narrow down the possible causes for this issue). In addition to this issue, there is another issue, which is related to the acquisition of interleaved slices, leading to this ‘zigzag’ effect. Note that it is possible to perform motion correction on interleaved scans and mitigate this effect, but it requires a bit more involvement from an image analysis standpoint. The idea is to split the EPI time series between odd and even slices, apply motion correction on each sub-volume, and then concatenate back the two corrected sub-volumes.
Hi, the results of running sct_check_dependencies are attached below.
And your point that the first video I attached and the file I sent you were from different subjects was correct. However, the subject of the file I sent you also had the same error as the video I am attaching.
Hope you can solve my problem.
Can you please install the latest SCT version, try again, and let us know how it goes?
Hi, I followed your advice and updated SCT to the latest version.
I have experienced the bug after sct_fmri_moco on two EPI images. And one of them was remedied by simply updating SCT to the latest version. However, I tried the same thing with the other one (the first video I sent when I started this thread) and he saw the same bug after sct_fmri_moco as before the SCT update.
The command used are as follows, as you wrote.
sct_fmri_moco -i fcn_rmvol_mcf_ref_SP.nii.gz -m mask_ref_SP_23.nii.gz
Can I send you (in a private email) two EPI images (before and after sct_fmri_moco) and a mask image of a subject that did not improve with the SCT update?
Hi, I have sent you the EPI image data before and after sct_fmri_moco, did you solve the problem?
Hi @elip97MGR,
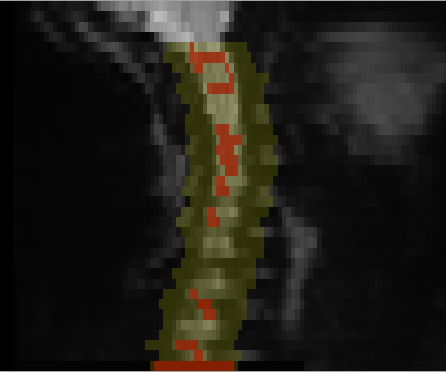
I have tried @jcohenadad’s “mask creation” + “cropping” steps from this comment on your most recent images (sent by email on Fri, Jan 12, 2024 – 369.9MB, dimensions [112, 112, 36, 236]):
Screenshots
However, I experienced the opposite problem – there is no motion correction at all. (moco_params.tsv → all X/Y translation are 0.0).
Looking at @jcohenadad’s commands, I notice that he both A) crops the image, and B) supplies a mask to the -m parameter of sct_fmri_moco. I think this is redundant – only one or the other is needed.
When, retrying sct_fmri_moco on just the cropped image (no -m), I do get motion correction  .
.
And, when I look at all of the volumes (using FSLeyes’ “movie” feature), I do not see any issues:
Screenshots

So, in short, I would recommend:
- Use @jcohenadad’s pipeline for mask creation as suggested here.
- Either crop the image using the mask, or supply the mask to
-m, but not both.
Please let me know if this fixes the issue on your end. 
Kind regards,
Joshua
1 Like