Hi!
- A description of the problem. (What are trying to do? What did you expect to happen? What actually happened?)
I’m trying to calculate the CSA of the spinal cord and the spinal canal using both segmentations (canal.nii.gz and cord.nii.gz), the vertebral label from totalspineseg and the same centerline (from the canal).
sct_process_segmentation -i canal.nii.gz -z 1:1684 -angle-corr-centerline canal.nii.gz -vertfile SPINE_110_step1_levels.nii.gz -perslice 1 -o csv_canal.csv
--
Spinal Cord Toolbox (6.5)
sct_process_segmentation -i canal.nii.gz -z 1:1684 -angle-corr-centerline canal.nii.gz -vertfile SPINE_110_step1_levels.nii.gz -perslice 1 -o csv_canal.csv
--
Converting image from type 'int16' to type 'float64' for linear interpolation
Converting image from type 'int16' to type 'float64' for linear interpolation
Compute shape analysis: 100%|█████████████| 1386/1386 [00:17<00:00, 81.48iter/s]
Aggregating metrics: 0%| | 0/9 [00:00<?, ?iter/s]/Users/fkmp/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/fromnumeric.py:3504: RuntimeWarning: Mean of empty slice.
return _methods._mean(a, axis=axis, dtype=dtype,
/Users/fkmp/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:129: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Aggregating metrics: 100%|██████████████████████| 9/9 [00:05<00:00, 1.72iter/s]
Done!
sct_process_segmentation -i cord.nii.gz -z 1:1684 -angle-corr-centerline canal.nii.gz -vertfile SPINE_110_step1_levels.nii.gz -perslice 1 -o csv_cord.csv
--
Spinal Cord Toolbox (6.5)
sct_process_segmentation -i cord.nii.gz -z 1:1684 -angle-corr-centerline canal.nii.gz -vertfile SPINE_110_step1_levels.nii.gz -perslice 1 -o csv_cord.csv
--
Converting image from type 'int16' to type 'float64' for linear interpolation
Converting image from type 'int16' to type 'float64' for linear interpolation
Compute shape analysis: 100%|████████████| 1063/1063 [00:07<00:00, 142.72iter/s]
Aggregating metrics: 0%| | 0/9 [00:00<?, ?iter/s]/Users/fkmp/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/fromnumeric.py:3504: RuntimeWarning: Mean of empty slice.
return _methods._mean(a, axis=axis, dtype=dtype,
/Users/fkmp/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:129: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Aggregating metrics: 100%|██████████████████████| 9/9 [00:05<00:00, 1.79iter/s]
Done!
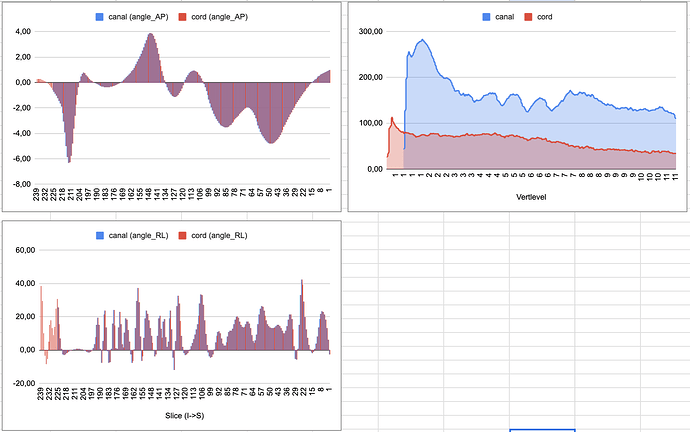
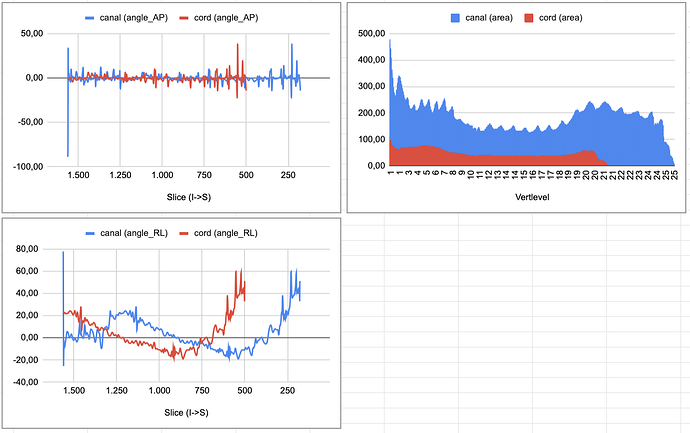
I obtained the results but then when I was verifying the correctness of the results, I realized that the angles are misplaced which I understand that implies that the results are not correct.
- System information. (Please run the command “sct_check_dependencies” in your terminal, then copy and paste the output.)
--
Spinal Cord Toolbox (6.5)
sct_check_dependencies
--
SYSTEM INFORMATION
------------------
SCT info:
- version: 6.5
- path: /Users/fkmp/sct_6.5
OS: osx (macOS-10.16-x86_64-i386-64bit)
CPU cores: Available: 8, Used by ITK functions: 8
RAM: Total: 8192MB, Used: 4867MB, Available: 2400MB
- File upload. (Please upload any data or scripts needed to reproduce your issue, if there are any.) →
canal.nii.gz (700.6 KB)
cord.nii.gz (562.1 KB)
SPINE_110_step1_levels.nii.gz (250.0 KB)