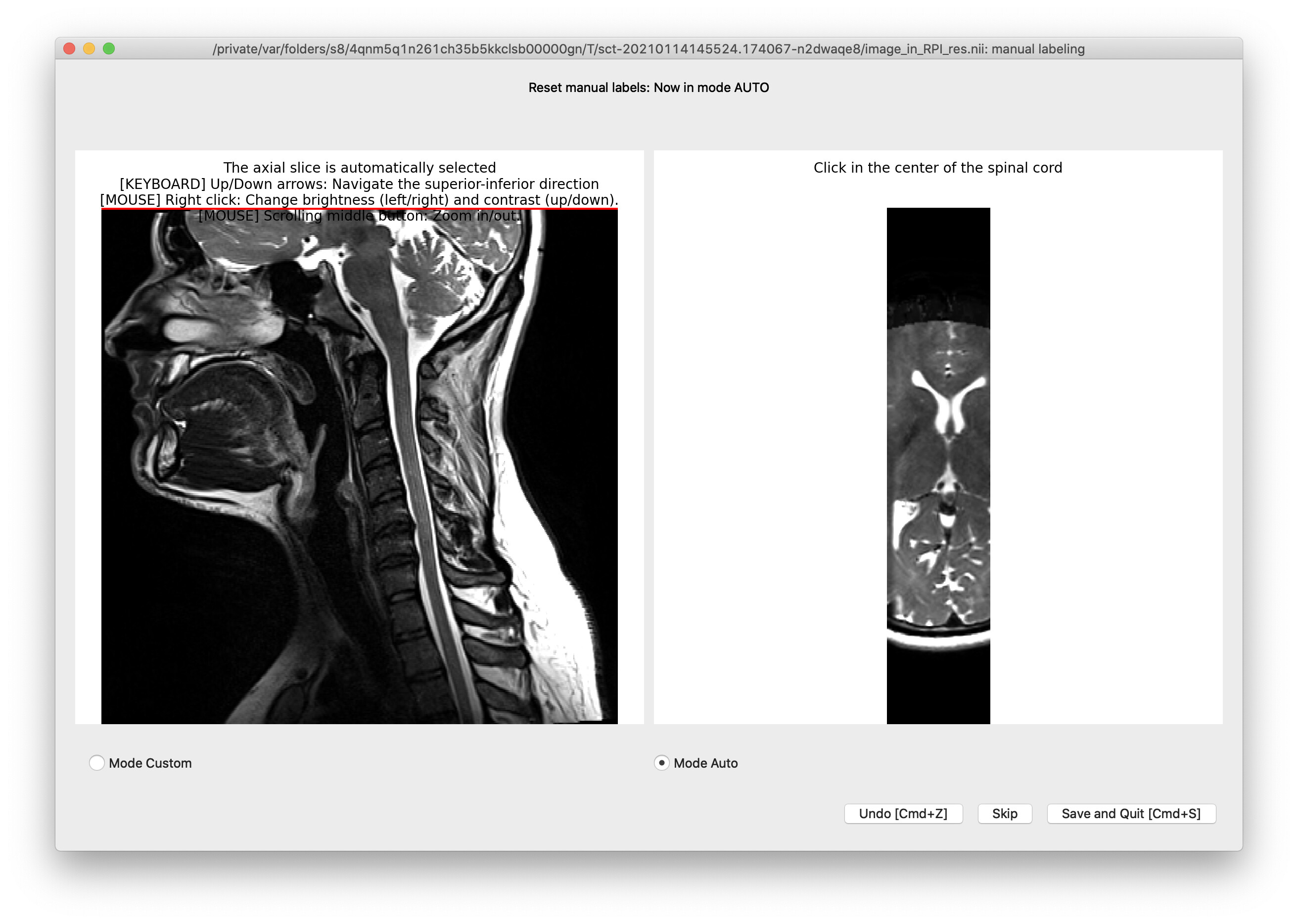
Hi @jcohenadad,
I am using the following command to help the SCT identify the thoracic spinal cord, which, as you know, can be more challenging in a handful of subjects:
sct_deepseg_sc -i t2.nii -c t2 -centerline viewer -brain 0 -kernel 2d
By default this viewer is in “Mode Auto”, which automatically initializes the first point a fair distance from the superior edge of the imaging volume (see above pic). I tried to fix this by switching to “Mode Custom”, but doing so then deletes(!) all the points defined during auto mode. And then if you define some point in custom mode and then switch back to auto mode, it deletes all the points again.
I have used this functionality a fair bit, and while it works well, I would like to suggest some simple potential enhancements:
- switching between custom mode and auto mode does not delete points previously defined.
- auto mode starts closer to the superior edge of the imaging volume… maybe a few mm from the top.
- if it could be helpful to the code in problematic (i.e, low SNR) regions(?), make the S-I step side in auto mode a parameter that the user can tweak (i.e., define 50% more control points).
The most important suggestion is #1 because doing a first pass in auto mode and then adding a few extra points in manual mode would be a terrifically awesome enhancement. 
Thank you!
-Rob
Hi Rob!
Thank you so much for the constructive feature requests. I’ve opened a ticket here where you can follow the implementation and chip in for further suggestions.
There is just one thing that I cannot reproduce, which is your second point. I’ve tried with a few images and the ‘auto’ mode always starts at the top slice. Eg:
To reproduce (v5.0.1):
sct_download_data -d sct_example_data
sct_deepseg_sc -i sct_example_data/t2/t2.nii.gz -c t2 -centerline viewer
If you confirm the unexpected behaviour on your end could you please upload (or DM me) the image?
Thanks!
Hi @jcohenadad - thanks for the quick reply! For these thoracic images, the first slice automatically selected in auto mode is always blank; therefore, I have to skip the first slice (there is no image to define the spinal cord on) and start with the second. I have been wondering if this has something to do with indexing the first slice or perhaps a rounding issue(?). I’m happy to share one of my de-identified data sets with you, if that could be helpful… -Rob
ahah! i suspect this is caused by gradient non-linearity correction, which slightly distorts the image (especially at the edge of the FOV), yielding edge slices (in all dimensions) with “almost zeroes” everywhere. So yes, I understand better the situation now.
I can see two workarounds:
- instead of showing a single axial slice, show the average of the 1-2 slices above and below;
- do not start at the first slice, but at the second (or third).
I would be more in favour of the first option.
Any other suggestion?
Hi @jcohenadad,
Ah! A brilliant idea. I downloaded the test data that you suggested above and do see the first slice, but I can’t recall seeing the first slice displayed in the viewer in any of the data sets that I’ve processed. Image warping caused by gradient non-linearity correction may indeed be the cause!
I agree that there are a couple potential solutions. I don’t know how widespread this issue is, but keeping things as they are but permit flexible switching between auto and manual mode should do the trick (I could define most of the points in auto mode and then go back to fix the top and bottom slices in manual mode). A more “elegant” solution might be to auto-detect if the first slice is all zeros, and if so start at the next non-zero slice (or average the current slice with the next slice).
Thank you! Please let me know if there’s anything that I can do to help. 
-Rob
1 Like
Hey Rob,
I’ve started drafting a pull request to incorporate the suggestions. If you have installed the git version of SCT, you can try the prototype branch by running:
cd $SCT_DIR
git checkout master
git pull
git checkout -b jca/centerline-viewer origin/jca/centerline-viewer
1 Like
Hi @jcohenadad,
Thank you for incorporating these suggestions so quickly! Wow.
I reprocessed 16 datasets this morning with your code from the new branch, and I’m pleased to report… < insert suspense here > … that the new centerline viewer code worked well in 100% of the subjects. Yay! 
One final comment that might be considered is how the “Undo”, “Delete all” and “Skip” buttons affect the sagittal and axial images. Currently, if any of these buttons are pressed, neither the sagittal nor axial images are immediately updated. For example, if I select “undo”, then I don’t see the point that I just “un-did” vanish; instead, it vanishes on the axial image only when I move the mouse cursor over it but still remains on the sagittal image until the next point is selected. Similarly, if I select “delete all”, then all the points remain on the sagittal image until the next point is selected – at which point all previous points vanish.
I am very pleased at the improved functionality of being able to switch back and forth between auto and manual mode because this will make the reprocessing of thoracic images much easier. THANK YOU!
-Rob
1 Like
Thank you for the feedback Rob!
We have now merged the PR, and will release 5.1.0 soon.
Other improvements and fixes will be addressed in subsequent releases. If you find something else, please feel free to contribute in this new issue.
Again, many thanks for your most valuable feedback!
Julien
1 Like
![]()