Hello SCT team,
I am having two sets of MRI images for the cervical spinal cord: T2 sagittal and T2 coronal. I am using T2 coronal to run the propseg, deepseg and other analysis. However, automatic and manual labelling of disc is very tough and imprecise as the associated sagittal view has low quality. However, the T2 sagittal view has clear images for disc and vertebral levels. Is there a way I can use T2 sagittal to get disc level info and use it to procced my analysis on T2 coronal sections.
I hope my question is clear.
Thank you,
Pawan
Hi @pawan_sharma,
Absolutely, you can perform vertebral (or disc) labeling on your sagittal image, and then bring the info to another orientation. See for example: Dwi metric computing - #4 by jcohenadad
@jcohenadad Thank you for guiding me to this resource. After running the recommended steps, I see a huge distortion in the multimodal registration. I am dealing with MRI of individuals with spinal cord injury and they may have missing vertebra or discs.
I see a huge distortion in the multimodal registration.
Without looking at the data it is very difficult for me to assess what is the best course of action. If you could share a representative example that would be helpful for me to guide you.
How can I share data with you? Is there any email id I can send it to?
Thank you,
Pawan
you can email me the data (my email is easy to find)
Thank you! I have shared the data with you on your university email id.
Pawan
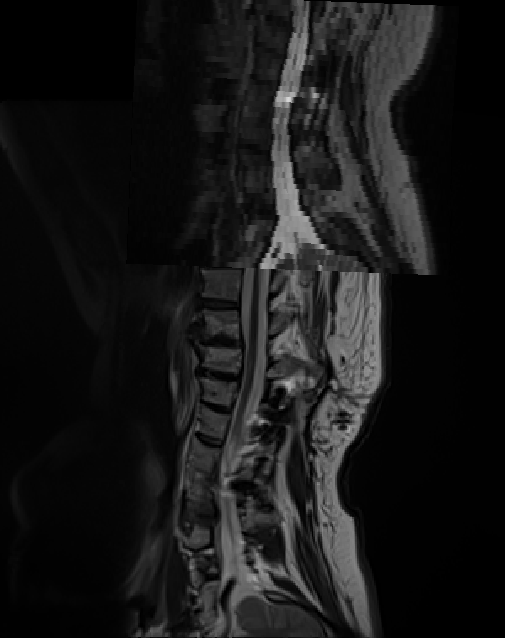
Hi @pawan_sharma , thank you for sending these two images.
I overlaid them on FSLeyes and I noticed that these two images are not defined within the same scanner coordinate system:
So, either there was a patient repositioning during the session, or these images were acquired across two different sessions, or the header of one of the NIfTI image is corrupted (maybe a DICOM to NIFTI conversion issue).
That makes total sense now. I will try using the recommended multi-modal registration for other patients and see if the same issue exists for their data.
Thank you,
Pawan
more importantly, before running the commands, I strongly recommend that you look at your images before doing any processing. Try to overlay them as I did. If you notice any problem there is no point in doing the analyses assuming images are aligned/ of sufficient quality.
That’s very useful suggestions. I would be doing that moving forward.
Thank you,
Pawan