Hello ?
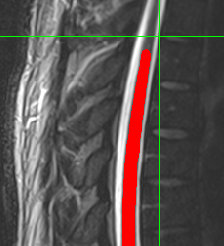
Segmentation result of conus region is not as good as C and T spinal cord. Therefore, I used different cord which was suggested in this forum.
sct_deepseg -i t2.nii.gz -task seg_lumbar_sc_t2w
The result was good.
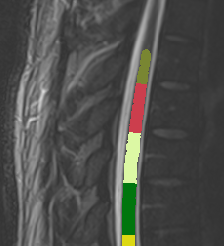
Then I want to calculate CSA of this result by normalizing PAM 50 template as follows ;
sct_process_segmentation -i t2_seg-manual.nii.gz -vertfile t2_seg-manual_labeled.nii.gz -perslice 1 -normalize-PAM50 1 -o csa_pam50_L.csv
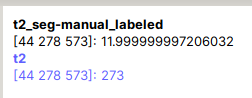
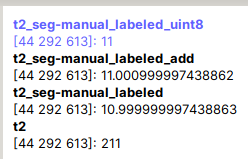
Following is error message
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Compute shape analysis: 100%|██████████████| 540/540 [00:05<00:00, 106.37iter/s]
Traceback (most recent call last):
File "/Users/ahn/sct_6.0/spinalcordtoolbox/scripts/sct_process_segmentation.py", line 521, in <module>
main(sys.argv[1:])
File "/Users/ahn/sct_6.0/spinalcordtoolbox/scripts/sct_process_segmentation.py", line 424, in main
metrics_PAM50_space = interpolate_metrics(metrics, fname_vert_level_PAM50, fname_vert_level)
File "/Users/ahn/sct_6.0/spinalcordtoolbox/metrics_to_PAM50.py", line 59, in interpolate_metrics
metrics_inter = np.interp(x_PAM50, x, metric_values_level)
File "<__array_function__ internals>", line 180, in interp
File "/Users/ahn/sct_6.0/python/envs/venv_sct/lib/python3.9/site-packages/numpy/lib/function_base.py", line 1594, in interp
return interp_func(x, xp, fp, left, right)
ValueError: array of sample points is empty
How can I solve this problem ?
Sincerely
Sung Jun Ahn
sct_check_dependencies
SYSTEM INFORMATION
------------------
SCT info:
- version: 6.0
- path: /Users/ahn/sct_6.0
OS: osx (macOS-10.16-x86_64-i386-64bit)
CPU cores: Available: 4, Used by ITK functions: 4
RAM: Total: 16384MB, Used: 6350MB, Available: 10032MB