Dear @jcohenadad!
Thank you very much for the clarifications.
To answer your question, our end goal is to have all of our subjects in a common space. We are acquiring these MEGRE scans under several conditions, and we want to compare signal intensities along the cord, across these conditions for all subjects, similar to the recent preprint by Kaptan et al.
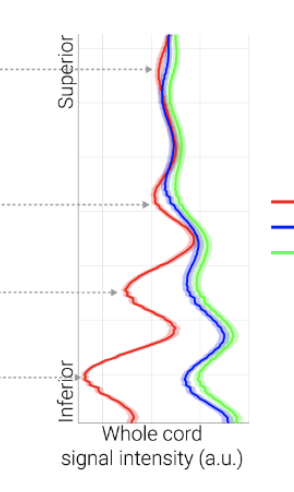
I have tried to go down the route of coregistering all subjects to a destination subject, but due to the poor quality of the MEGRE scans, that requires applying three warps, using the T1s as an in-between (source MEGRE to source T1; source T1 to destination T1; destination T1 to destination source). Those results did not satisfy me, and I am exploring alternatives.
I much prefer going down your suggested route of PAM50 registration
For comparison, here are the two pipelines for PAM50 regiistration:
Direct coregistration to ME-GRE to PAM50
This pipeline is almost identical to your proposal, only I switched “-x linear” to “-x nn” in the step where the segmentation is warped back to the ME-GRE. Trilinear interpolation gives non-binary values in the segmentation, see Note 1 on the bottom.
# Segment cord on T1w
sct_deepseg_sc -i Lowres_run1_T1w.nii.gz -c t1 -qc qc
# Generate mask around the cord (for subsequent registration)
sct_create_mask -i Lowres_run1_T1w.nii.gz -p centerline,Lowres_run1_T1w_seg.nii.gz -size 35mm -f cylinder
# Register the T1w scan to the mean ME-GRE scan
# Tips: Here we specify '-dseg Lowres_run1_T1w_seg.nii.gz' only to be able to generate the QC report. Even thought 'Lowres_run1_T1w_seg.nii.gz' corresponds to iseg (not dseg), we don't really care because the segmentation is *not* used for registration (it is only used to center the spinal cord during QC).
sct_register_multimodal -i Lowres_run1_T1w.nii.gz -d Lowres_GRE_mean.nii.gz -dseg Lowres_run1_T1w_seg.nii.gz -m mask_Lowres_run1_T1w.nii.gz -param step=1,type=im,algo=slicereg,metric=CC -qc qc
# Apply the transformation to the cord segmentation
# Tips: we use trilinear interpolation because having the 'soft' segmentation will provide more accurate registration to the template
sct_apply_transfo -i Lowres_run1_T1w_seg.nii.gz -d Lowres_GRE_mean.nii.gz -w warp_Lowres_run1_T1w2Lowres_GRE_mean.nii.gz -x linear
# Create label centered in the cord at level C6/C7
sct_label_utils -i Lowres_run1_T1w_seg_reg.nii.gz -create-seg-mid 7 -o label_c6c7.nii.gz -qc qc
# Register the ME-GRE scan to the PAM50 template
# Tips: we use '-ref subject' because we don't want straightening (due to the thick slices).
# Tips: we only use slice-wise rigid translation, based on the segmentation, because due to the low resolution of the ME-GRE image, any non-affine registration would be 'risky'.
sct_register_to_template -i Lowres_GRE_mean.nii.gz -s Lowres_run1_T1w_seg_reg.nii.gz -ldisc label_c6c7.nii.gz -ref subject -param step=1,type=seg,algo=centermassrot -qc qc
Using the T1w as an intermediary step
# Segment cord on T1w
sct_deepseg_sc -i Lowres_run1_T1w.nii.gz -c t1 -qc qc
# Generate mask around the cord (for subsequent registration)
sct_create_mask -i Lowres_run1_T1w.nii.gz -p centerline,Lowres_run1_T1w_seg.nii.gz -size 35mm -f cylinder
# Register the T1w scan to the mean ME-GRE scan
# Tips: Here we specify '-dseg Lowres_run1_T1w_seg.nii.gz' only to be able to generate the QC report. Even thought 'Lowres_run1_T1w_seg.nii.gz' corresponds to iseg (not dseg), we don't really care because the segmentation is *not* used for registration (it is only used to center the spinal cord during QC).
sct_register_multimodal -i Lowres_run1_T1w.nii.gz -d Lowres_GRE_mean.nii.gz -dseg Lowres_run1_T1w_seg.nii.gz -m mask_Lowres_run1_T1w.nii.gz -param step=1,type=im,algo=slicereg,metric=CC -qc qc
#New steps start here
# Create label centered in the cord at level C6/C7 on the T1w!
sct_label_utils -i Lowres_run1_T1w_seg.nii.gz -create-seg-mid 7 -o label_c6c7.nii.gz -qc qc
# Coregistering the T1w to the PAM50. I am using your proposed one-step, slicewise method
sct_register_to_template -i Lowres_run1_T1w.nii.gz -s Lowres_run1_T1w_seg.nii.gz -ldisc label_c6c7.nii.gz -ref subject -param step=1,type=seg,algo=centermassrot -qc qc
# Concatenating the warp fields
sct_concat_transfo -d /Users/danielpapp/spinalcordtoolbox/data/PAM50/template/PAM50_t1.nii.gz -w warp_Lowres_GRE_mean2Lowres_run1_T1w.nii.gz warp_anat2template.nii.gz -o warp_MEGRE_2_PAM50.nii.gz
# Finally, applying the concatinated warp field to the MEGRE
sct_apply_transfo -d /Users/danielpapp/spinalcordtoolbox/data/PAM50/template/PAM50_t1.nii.gz -w warp_MEGRE_2_PAM50.nii.gz -i Lowres_GRE_mean.nii.gz
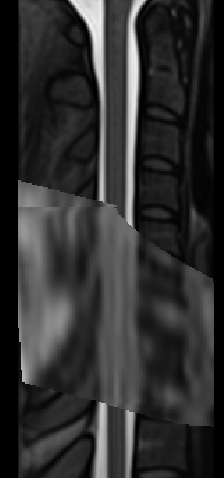
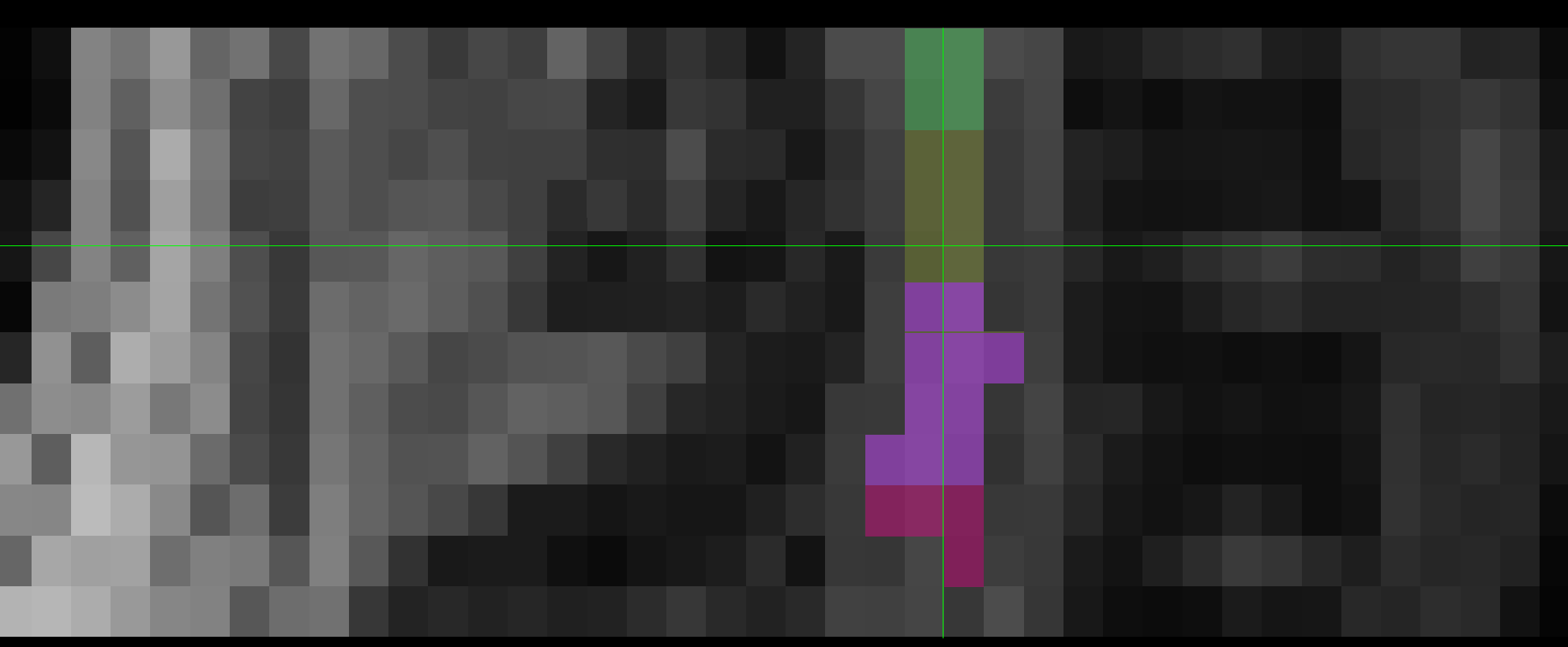
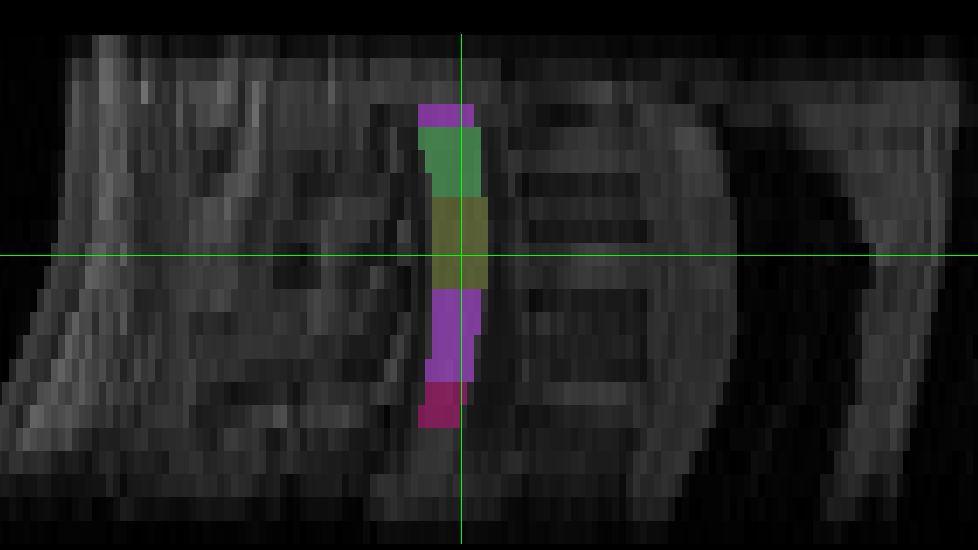
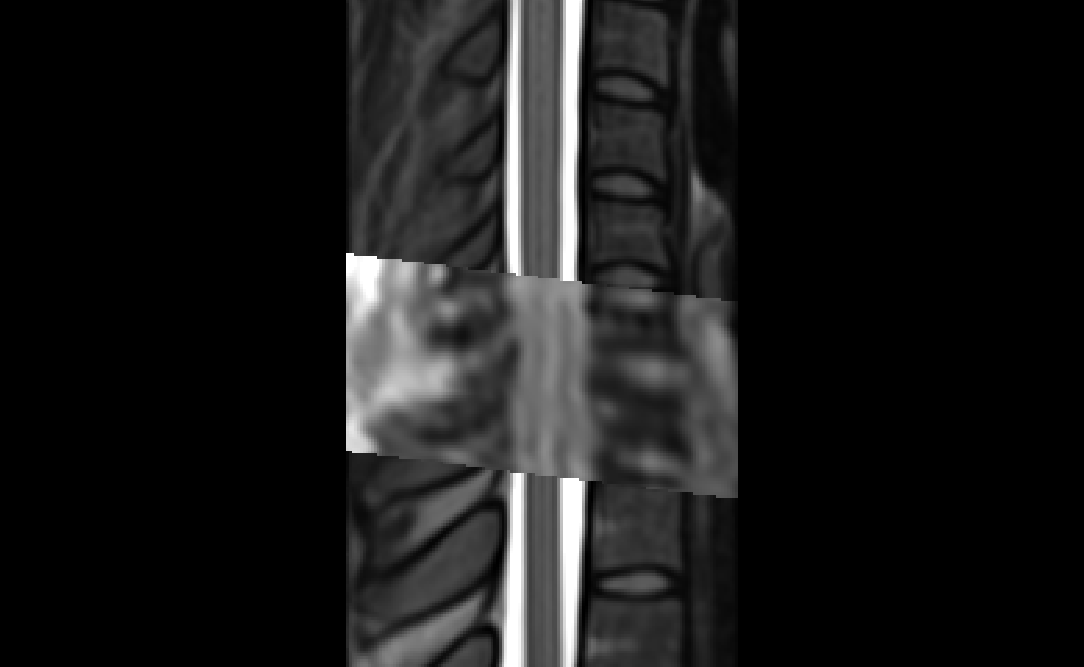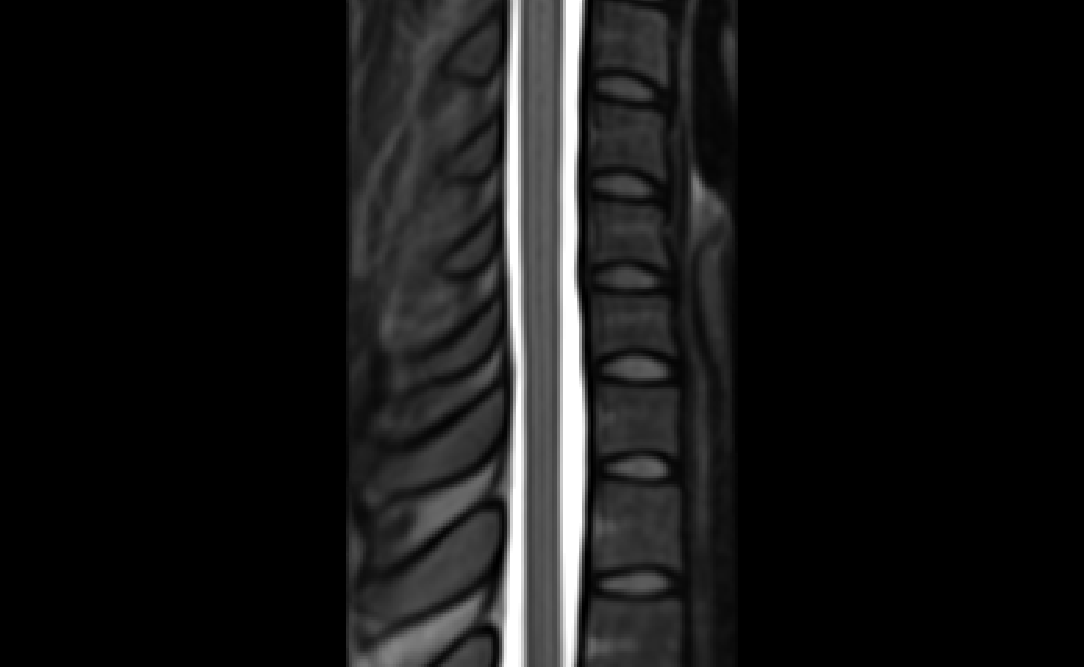
The two results are quite dissimilar. The direct coregistration seems stretched in the Z direction, compared to the 2-step process, and there is, for lack of a better word, a wavy, sinusoidal nature to the saggital view. The gif loops as PAM50–>direct coregistration–>2step coregistration
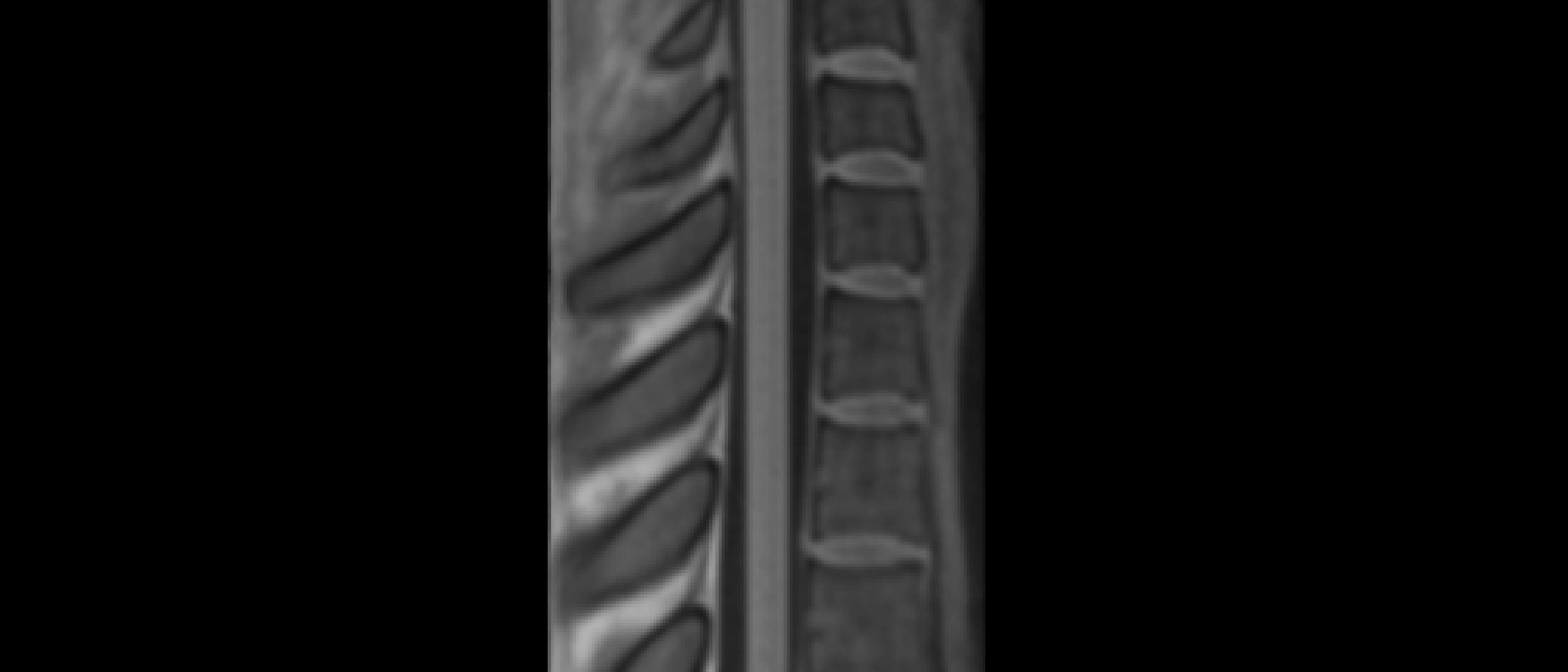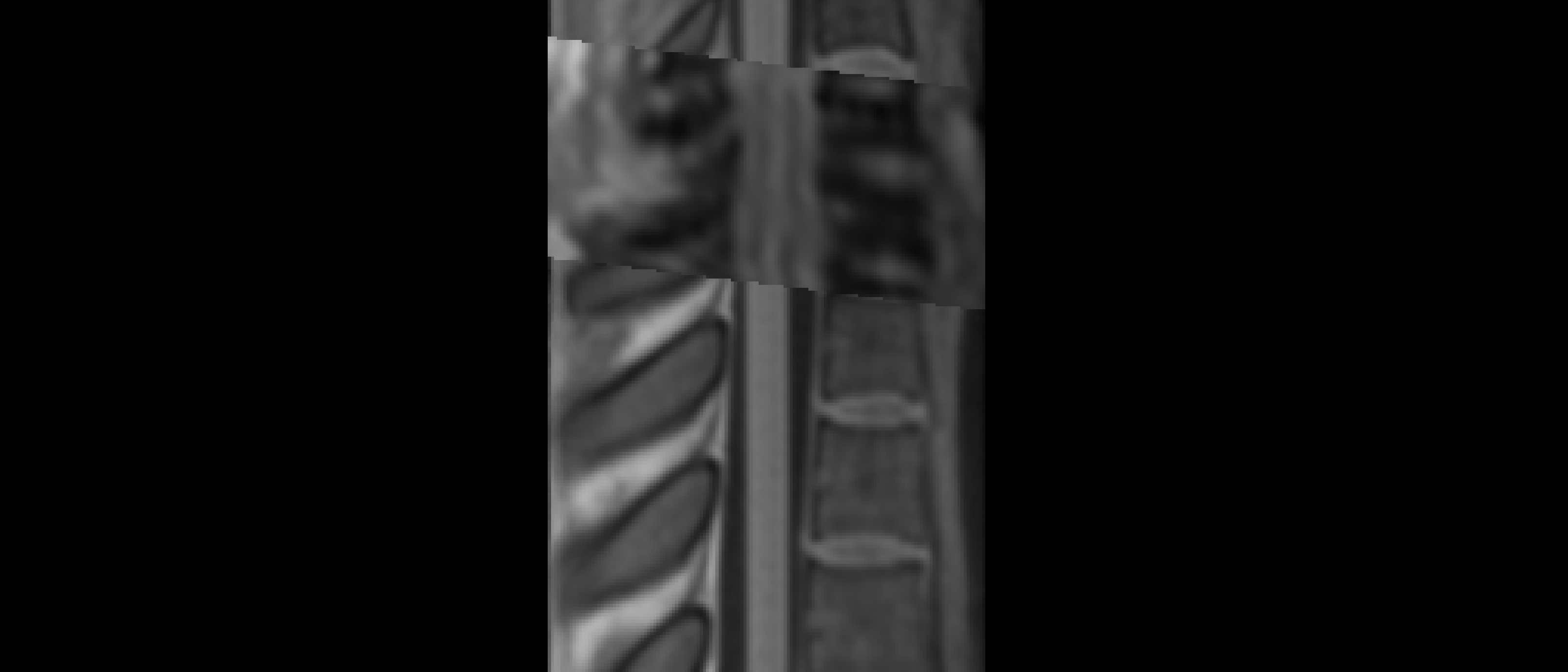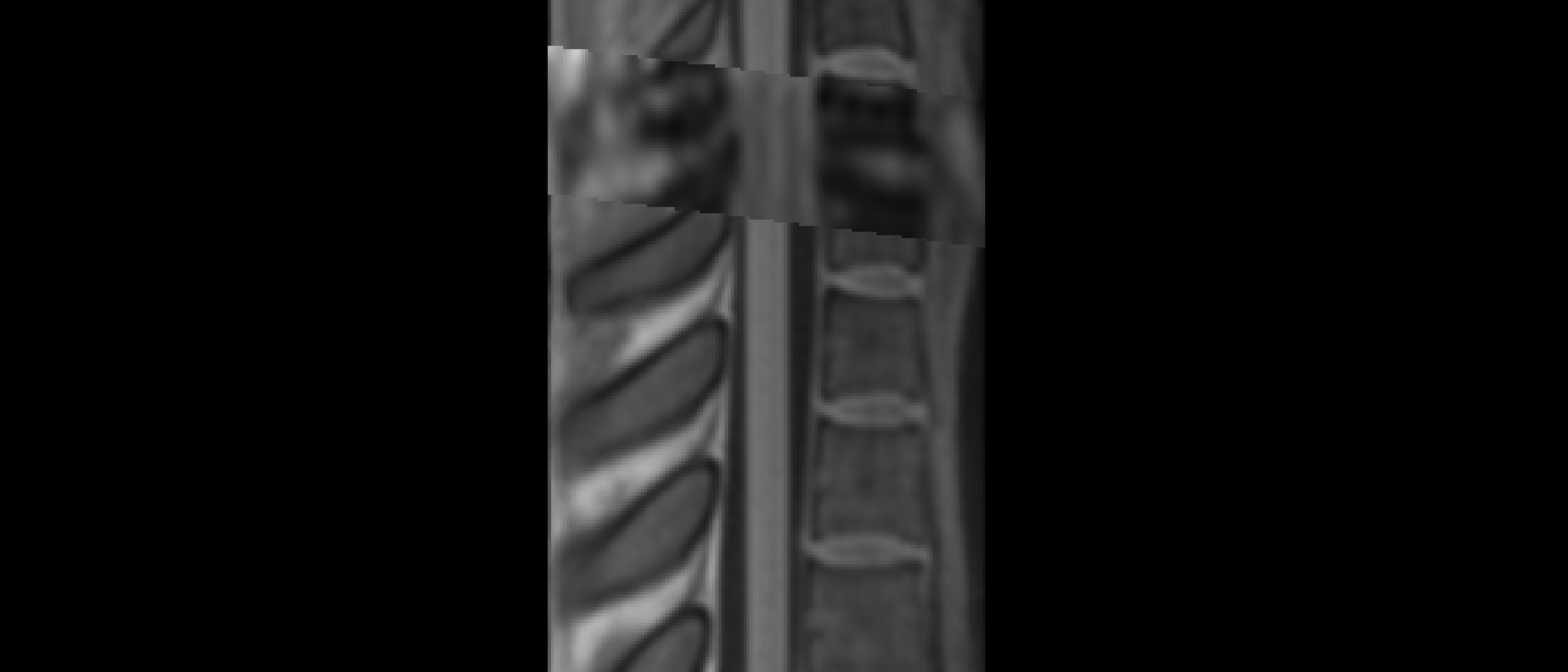
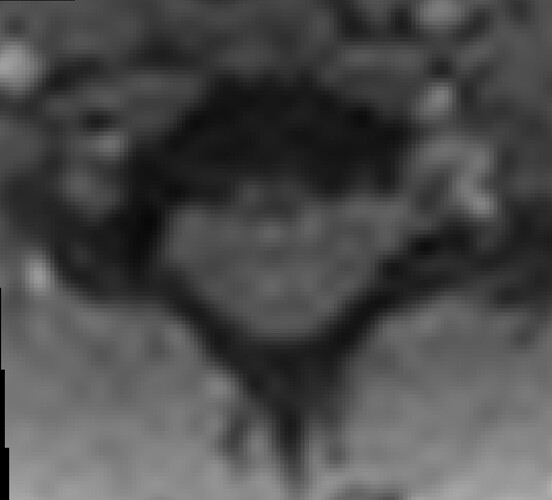
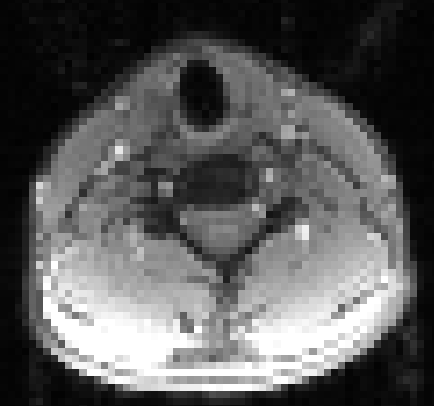
In the axial view, there is a rotation in the direct coregistration that is missing (I assume this is a good thing) in the 2step coregisration. Gif loops the same as previous. The images still look interpolated, but I guess that is just the result of the low resolution for the initial input.
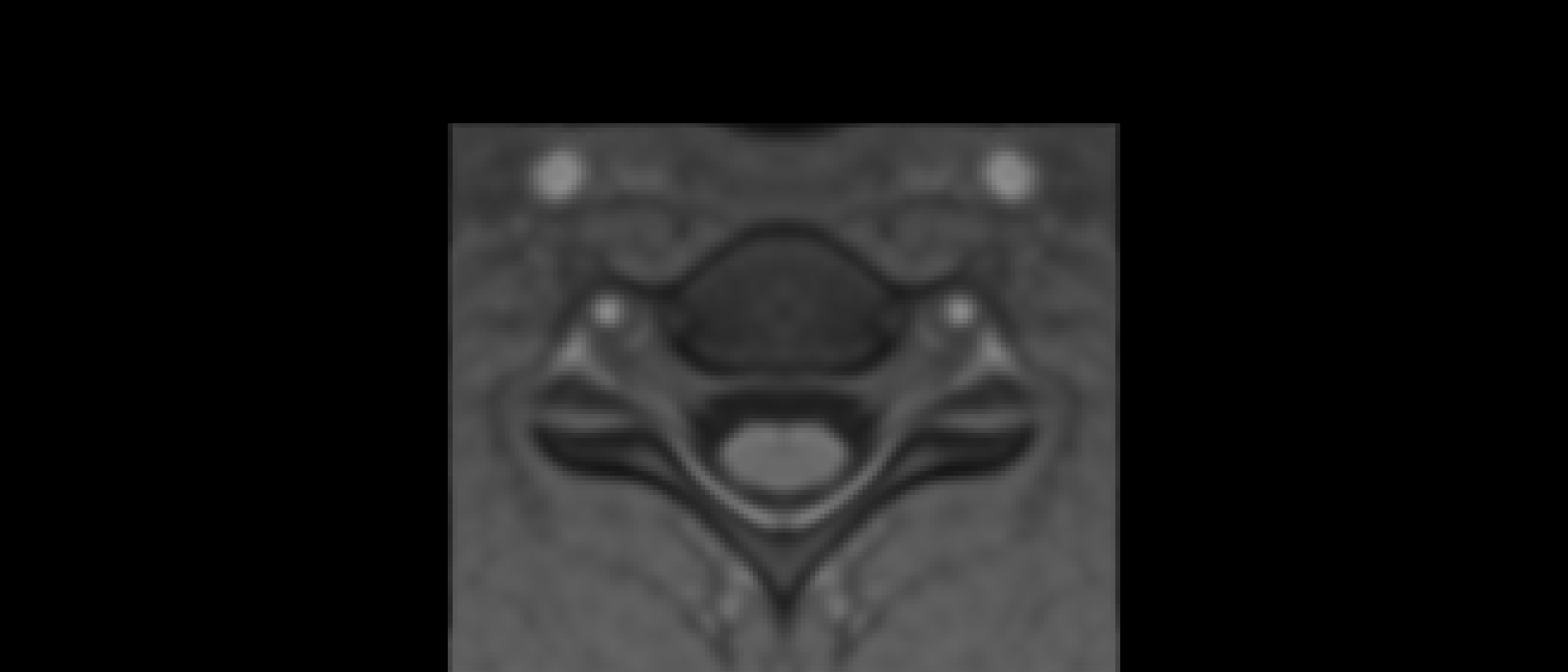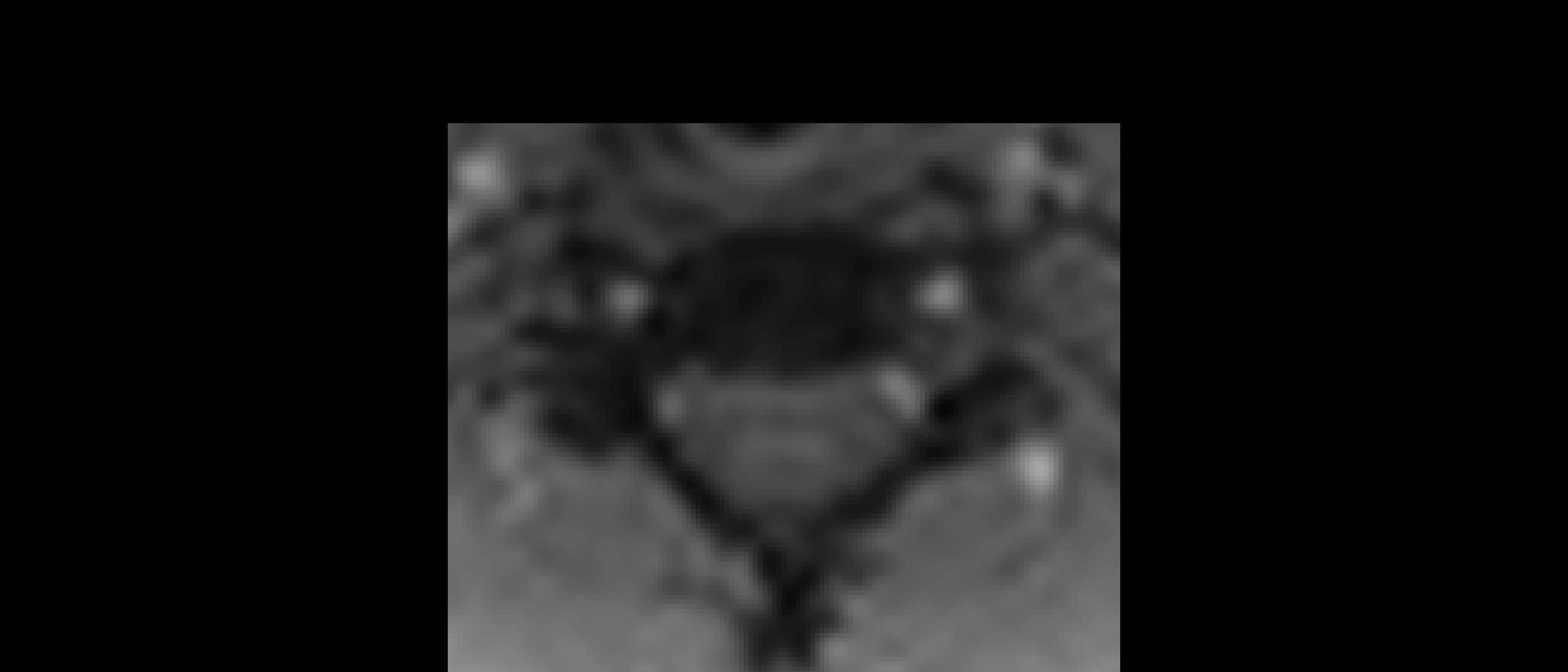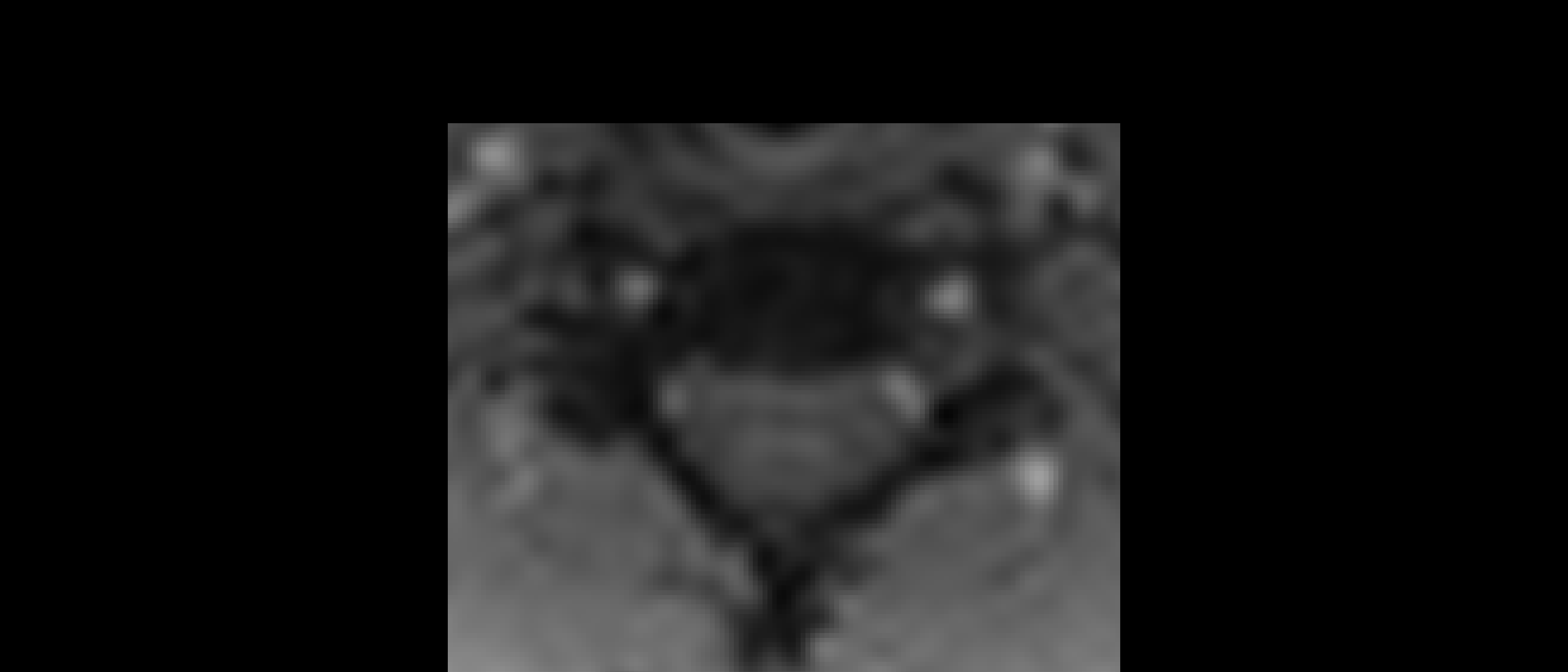
I will stick with the 2-step registration to PAM50, based on these results.
Note 1:
Trilinear interpolation on the spinal cord segmentation results in non-binary values within the warped segmentation. I do not know how SCT deals with this. Naively, I would assume that non-binary values are ignored.
Note 2: The files resulting from coregistration to PAM50 are saved as 64-bit float, I assume to accomodate negative floating point values that result from coregistration. Is there a way to tell SCT during the coreg step to save the data as INT16 (which is enough for our purposes?)
I have been using the following hack, but I don’t know if sct_register_to_template has a hidden “-type” flag too
sct_maths -i anat2template.nii.gz -o anat2template_16.nii.gz -add 0 -type int16
Thanks!
Daniel