Dear SCT hivemind!
I have a potential issue with coregistering some scans to PAM50. I have a long series of 2DGRE scans of the C-spine scans that I want to coregister to the PAM50 template in an automated way. I tried a couple of pipelines, and I would like your input in which should be best. I also have a 3D MPRAGE scan of the same FOV, that I use to help in some of the pipelines.
My preferred outcome is an automated pipeline that will deliver good coregistration to the PAM50 template from the C2/C3 to the T2/T3 levels. My issue is that sometimes, like on the data set attached to this post, the automatic detection of the C2/C3 disk fails when using sct_label_vertebrate, and I have to work around this. I have not found any fully satisfying solution.
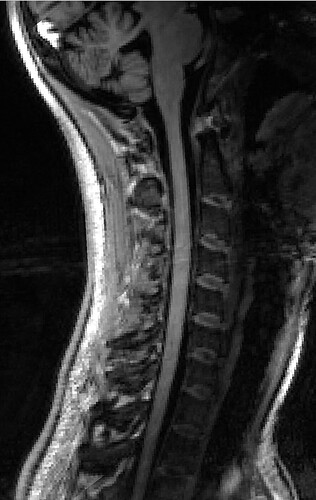
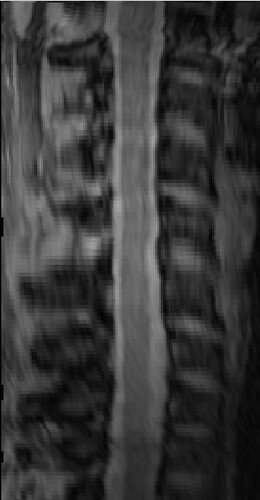
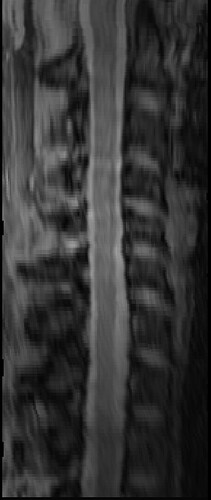
The overall image quality is like this:
GRE scan
MPRAGE scan
Pipeline 1: Using only the 2D images
Here, I am forced to initialize the sct_label_vertebrate funtion with some position information, but I have found it quite susceptible to the exact choice of this initz label.
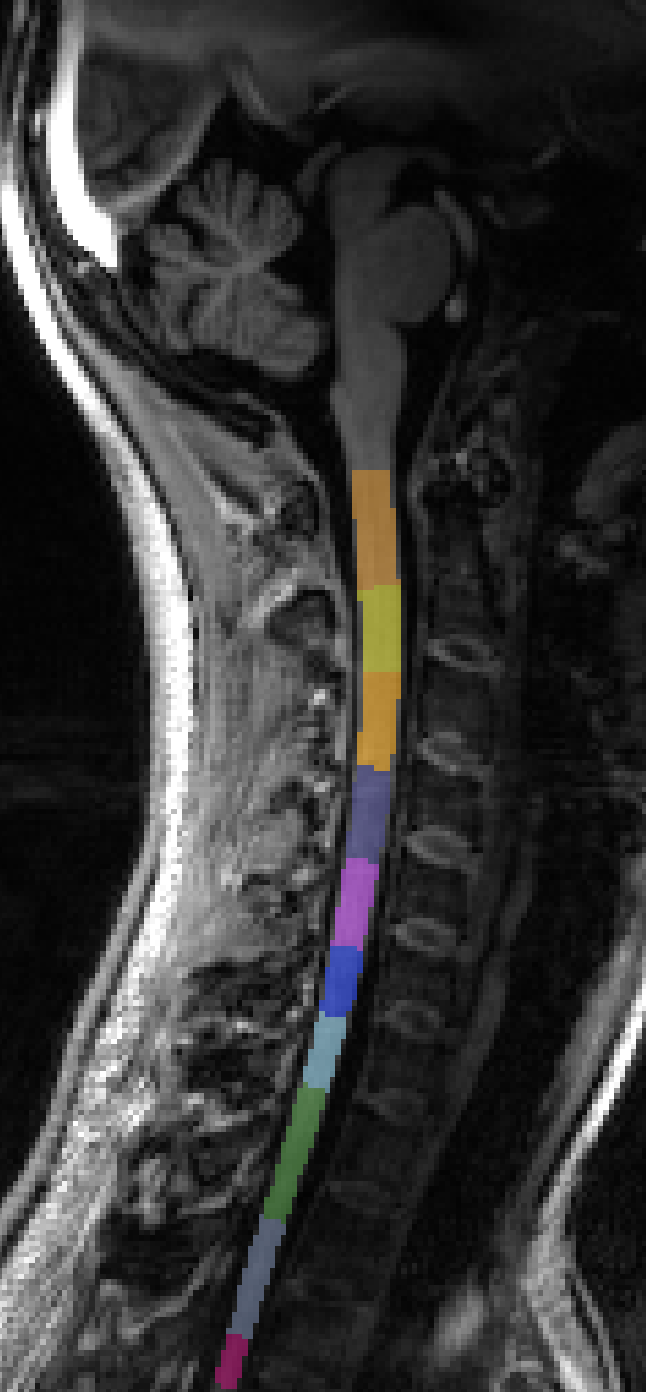
Initializing at C2/C3 results in the fourth label being centered on the C3/C4 disk, and then everything is a bit squashed.
sct_label_vertebrae -i 22_gre2d_06mm_FOV230_70sli_VolSpec.nii.gz -s 22_gre2d_06mm_FOV230_70sli_VolSpec_seg.nii.gz -c t2 -initz 44,3
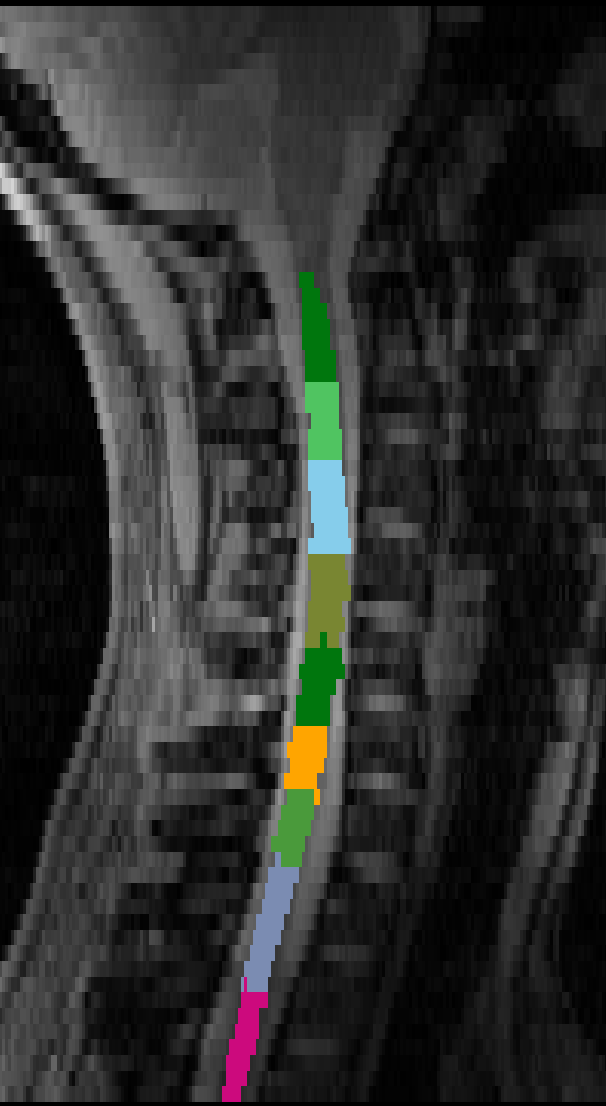
Initialising with C4/C5 stretches the second label too much down the body of C3:
sct_label_vertebrae -i 22_gre2d_06mm_FOV230_70sli_VolSpec.nii.gz -s 22_gre2d_06mm_FOV230_70sli_VolSpec_seg.nii.gz -c t2 -initz 31,5
As a result, the vertebral disk labels that I use for coregistration are a bit off, this seems to cause the coregistration around C5-C7 to be suboptimal:
sct_register_to_template -i 22_gre2d_06mm_FOV230_70sli_VolSpec.nii.gz -s 22_gre2d_06mm_FOV230_70sli_VolSpec_seg.nii.gz -ldisc 22_gre2d_06mm_FOV230_70sli_VolSpec_seg_labeled_discs_initzC5.nii.gz
I know of the alternative way of extracting some disk labels from the label file, but that would be another source of human variability and error, something I’d like to avoid.
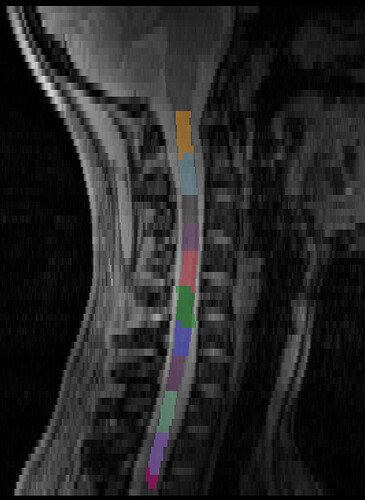
Pipeline 2: Utilizing the MPRAGE
Since I have the MPRAGE scan available, and since the automatic labeling works quite reliably, at least in the C2-C7 range, I want to transfer the label from one scan to the other.
I coregister the MPRAGE to the GRE:
sct_register_multimodal -i 47_t1_mprage_sag_p2_1mm_noRFshim.nii.gz -iseg 47_t1_mprage_sag_p2_1mm_noRFshim_seg.nii.gz -d 22_gre2d_06mm_FOV230_70sli_VolSpec.nii.gz -dseg 22_gre2d_06mm_FOV230_70sli_VolSpec_seg.nii.gz
Apply this warp to the label file:
sct_apply_transfo -i 47_t1_mprage_sag_p2_1mm_noRFshim_seg_labeled.nii.gz -d 22_gre2d_06mm_FOV230_70sli_VolSpec.nii.gz -w warp_47_t1_mprage_sag_p2_1mm_noRFshim222_gre2d_06mm_FOV230_70sli_VolSpec.nii.gz -x nn
This looks decent, but may be mislabeling around C6/C7?
Finally I extract the midbody labels from this warped label file:
sct_label_utils -i 47_t1_mprage_sag_p2_1mm_noRFshim_seg_labeled_reg.nii.gz -vert-body 0
And use these new labels to help with the coregistration to PAM50:
sct_register_to_template -i 22_gre2d_06mm_FOV230_70sli_VolSpec.nii.gz -s 22_gre2d_06mm_FOV230_70sli_VolSpec_seg.nii.gz -lspinal labels.nii.gz -c t2
This, sadly, also looks quite bad around C5-C6:
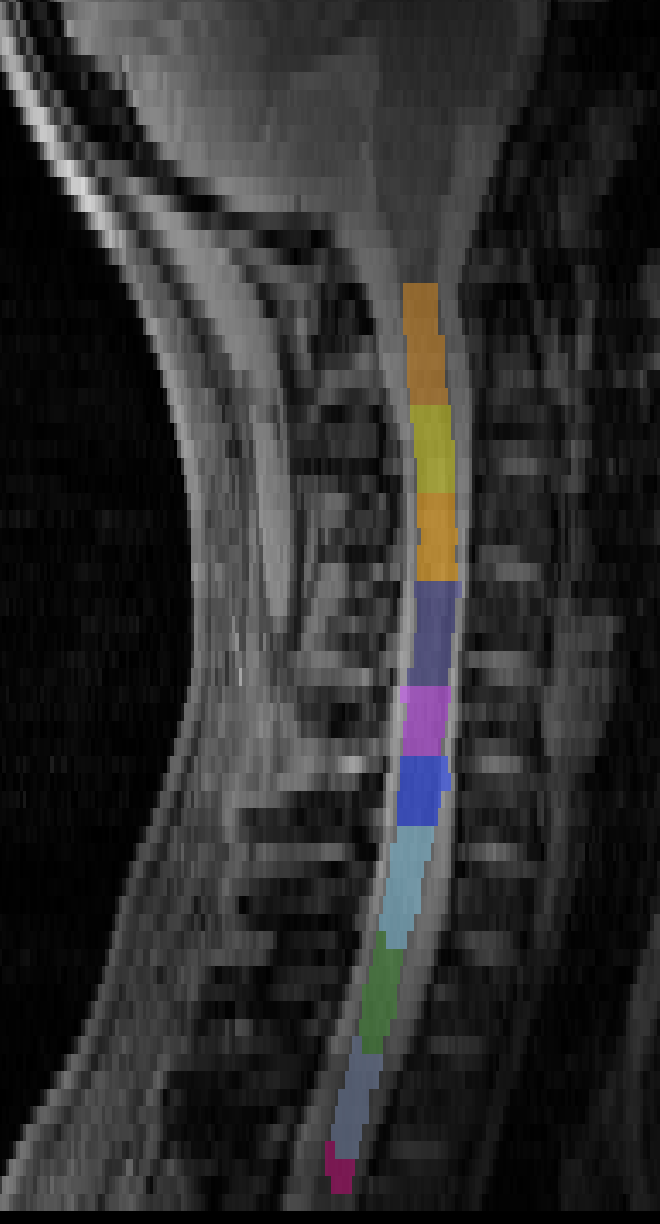
Pipeline 3: Manual labeling from C3 to T2
I assumed the best way to solve this would be to manually label everything:
sct_label_utils -i 22_gre2d_06mm_FOV230_70sli_VolSpec.nii.gz -create-viewer 1,2,3,4,5,6,7,8,9,10
The result is the best so far, but can’t be automated.
Is there a way to do this labeling and coregistration in an automated way even when the image quality is poor, or does there need to be human supervision and fixing? I hope I am just being paranoid and overly cautious, but I value expert input.
SCT version:
sct_version
git-master-ec93f822cd032c08a70026113d88b8f8e0438392
Pulled fresh last morning at time of writing (21.11.2023)