Hi SCT Team,
I am following along with yesterday’s class slides and I am getting a clipped version of the template when I use sct_warp_template. This causes me to loose my lower level for my vertebral labels from the template. Is there a reason that the CSA calculations are done on the registered template levels instead of the levels that are calculated with sct_label_vertebrae?
Also, is there a way to get all of the template registered into the subject space, not just a limited FOV?
Blake
Hi Blake,
I am getting a clipped version of the template when I use sct_warp_template
how much clipping? Have you used more than 2 labels? If so, you might want to label also the parts that you don’t want to see clipped.
Is there a reason that the CSA calculations are done on the registered template levels instead of the levels that are calculated with sct_label_vertebrae
Only a practical reason, because in most cases the registered template covers the region of interest. But yes, you could also use an external labeling:
sct_process_segmentation -i SEG -vertfile SEG_LABELED
Also, is there a way to get all of the template registered into the subject space, not just a limited FOV?
yes, let’s see what’s happening in your case
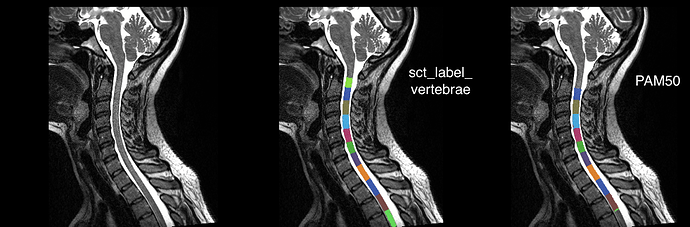
Here are my sample results. For the registration, I used the -ldisc option using the automated discs image from sct_label_vertebrae.
when inputting more than 2 labels (which is your case), the disc-alignment algorithm which is part of sct_straighten_spinalcord (called by sct_register_to_template) will crop the template for Z values that are outside the most upper and lower labels. This has to be done because wiht more than 3 labels, a non-linear fitting is performed along the superior-inferior axis, and there is no easy way to extrapolate this scaling factor if there are no more labels. So, to avoid that problem, you could either:
- limit the number of labels to 2 (use
sct_label_utils to select two labels at, let’s say, C2 and T3), as exemplified here, or
- manually add disc labels at the top of C1 as well as at the most lower part of you labeled segmentation. I do not recommend that approach however, because in this case, it “seems” like that bottom of your labeled segmentation coincides with T4-T5 disc, so the straightening will align this disc with the PAM50 T4-T5 disc. But if in other subjects, the FOV stops in the middle of the vertebral body, the “apparent” location of the T4-T5 disc will be wrong in your subject and that error will be reflected when registering with the PAM50 template.
So, while the first approach seems more “crude” in terms of disc alignment, it generalizes better across your population, and also keep in mind that vertebral-based labeling is only a crude approximate of the actual spinal levels, so I would not worry to much if the registered discs are a few mm away from the PAM50 template between C2 and T3 (if you decide to go with these levels for labeling).