Hello guys!
I am running CSA analysis for different studies, but recently I came across with a potential issue; when I analyze some nii the majority of the CSA in the cervical spinal cord are in magnitude of 10s (ie C3 45) but with one study I am finding magnitudes of 100 (ie 900). Can someone give me some orientation of what is happening?
Hi @enriquegf,
Thank you for reaching out. Could you please clarify what you mean by “10s (ie C3 45)”? CSA are in mm^2, so I am not sure what you meant here.
I’m also confused by “magnitudes of 100 (ie 900)”. What is the unit here? And what does “ie 900” mean?
I am sorry for this barely legible explanation. I upload both screenshot from what I ve done.
I obtained results from one study in magnitude of hundreds, but in the same patient in a previous study it is in magnitude of tens.
It is probably caused by your NIfTI headers, which are corrupted (ie: the qform/sform does not reflect the actual physical dimensions). If you upload an example image and segmentation that yields those CSAs I can have a look.
ok! here is the .nii of this patient from which I obtained this results.
Thank very very much! I am just beginning to try SCT as a neurologist m20.nii.gz (2.4 MB)
hum, the header looks good-- could you please also upload the segmentation you used to compute the CSA?
Of course! here it is
m20_seg_labeled_discs.nii.gz (77.7 KB) m20_seg_labeled.nii.gz (84.3 KB) m20_seg.nii.gz (4.8 KB)
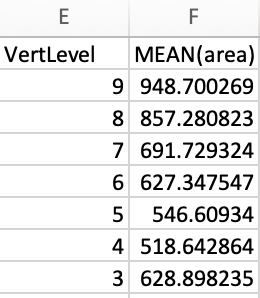
I’m not sure what you did, but results look OK on my end:
| VertLevel | MEAN(area) |
|---|---|
| 8 | 44.05825885019590 |
| 7 | 48.594102619341700 |
| 6 | 53.87744611037150 |
| 5 | 63.68126638238550 |
| 4 | 57.07686534077420 |
| 3 | 57.532144223958000 |
CSV output: csa.csv (2.9 KB)
I used this command (SCT v5.1.0):
sct_process_segmentation -i m20_seg.nii.gz -vertfile m20_seg_labeled.nii.gz -vert 3:8 -perlevel 1
Thanks!
I will re-run it to try to look for what error I might have.
If you could write the exact syntax you typed it is helpful for us to debug. Usually we ask users to copy/paste the syntax and output from the Terminal.
no news from the user after one month so I’m tagging this issue as “solved”
I am sorry for the delay Prof. Cohen,
I have been re running this same analysis and found again the same situation. In this occasion I will paste the text:
MacBook-QQ:t2_tse_sag_320_16 enriquegomezfigueroa$ sct_propseg -i m21.nii.gz -c t2
–
Spinal Cord Toolbox (4.3)
Create temporary folder (/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314112256.300851-label_vertebrae-0pqrzedw)…
Remove temporary files…
rm -rf /var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314112256.305088-7059gwqu
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_propseg -t t2 -o ./ -verbose -i ‘/Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16/m21.nii.gz’ -init-centerline /var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314112256.300851-label_vertebrae-0pqrzedw/centerline_optic.nii.gz -centerline-binary # in /Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16
Check consistency of segmentation…
Create temporary folder (/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314112326.853661-propseg-ym5n60am)…
tmp.segmentation.nii.gz
tmp.centerline.nii.gz
Get data dimensions…
usage: sct_image -i [ …] [-h] [-o ] [-pad ]
[-pad-asym ] [-split {x,y,z,t}] [-concat {x,y,z,t}]
[-remove-vol ] [-keep-vol ]
[-type {uint8,int16,int32,float32,complex64,float64,int8,uint16,uint32,int64,uint64}]
[-copy-header ] [-getorient]
[-setorient {RIP,LIP,RSP,LSP,RIA,LIA,RSA,LSA,IRP,ILP,SRP,SLP,IRA,ILA,SRA,SLA,RPI,LPI,RAI,LAI,RPS,LPS,RAS,LAS,PRI,PLI,ARI,ALI,PRS,PLS,ARS,ALS,IPR,SPR,IAR,SAR,IPL,SPL,IAL,SAL,PIR,PSR,AIR,ASR,PIL,PSL,AIL,ASL}]
[-setorient-data {RIP,LIP,RSP,LSP,RIA,LIA,RSA,LSA,IRP,ILP,SRP,SLP,IRA,ILA,SRA,SLA,RPI,LPI,RAI,LAI,RPS,LPS,RAS,LAS,PRI,PLI,ARI,ALI,PRS,PLS,ARS,ALS,IPR,SPR,IAR,SAR,IPL,SPL,IAL,SAL,PIR,PSR,AIR,ASR,PIL,PSL,AIL,ASL}]
[-mcs] [-omc] [-display-warp] [-to-fsl [ [ …]]]
[-v {0,1,2}]
sct_image: error: unrecognized arguments: Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16/m21_seg.nii.gz
MacBook-QQ:t2_tse_sag_320_16 enriquegomezfigueroa$ sct_label_vertebrae -i m21.nii.gz -s m21_seg.nii.gz -c t2
–
Spinal Cord Toolbox (4.3)
Create temporary folder (/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314113350.857435-label_vertebrae-ti9i761j)…
Copying input data to tmp folder…
Straighten spinal cord…
Create temporary folder (/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314113350.960104-straighten_spinalcord-14jub3zi)…
Copy files to tmp folder…
Window length needs to be >= 3. Returning input signal.
Window length needs to be >= 3. Returning input signal.
Fitting centerline using B-spline approximation
Error on approximation = 24.28 mm
Error on approximation = 5.47 mm
Error on approximation = 5.46 mm
Error on approximation = 2.85 mm
Error on approximation = 1.25 mm
Error on approximation = 0.56 mm
Error on approximation = 0.36 mm
Error on approximation = 0.38 mm
Error on approximation = 0.42 mm
Error on approximation = 0.32 mm
Error on approximation = 0.22 mm
Error on approximation = 0.22 mm
Error on approximation = 0.24 mm
Error on approximation = 0.19 mm
Error on approximation = 0.19 mm
Error on approximation = 0.2 mm
Error on approximation = 0.2 mm
Error on approximation = 0.17 mm
Error on approximation = 0.17 mm
Error on approximation = 0.17 mm
Error on approximation = 0.16 mm
Error on approximation = 0.1 mm
Error on approximation = 0.06 mm
Error on approximation = 0.06 mm
Error on approximation = 0.09 mm
Error on approximation = 0.09 mm
Error on approximation = 0.07 mm
Error on approximation = 0.06 mm
Error on approximation = 0.05 mm
Error on approximation = 0.05 mm
Error on approximation = 0.05 mm
Create the straight space and the safe zone
Length of spinal cord: 235.5724424780292
Size of spinal cord in z direction: 219.3125
Ratio length/size: 1.0741405185661064
Safe zone boundaries (curved space): [-157.9805145263672, 61.33198547363281]
Safe zone boundaries (straight space): [-166.11048576538178, 69.46195671264742]
Pad input volume to account for spinal cord length…
Time to generate centerline: 41.0 ms
100%|████████████████████████████████████████| 344/344 [00:01<00:00, 190.79it/s]
100%|█████████████████████████████████████████| 356/356 [00:09<00:00, 37.02it/s]
Warping field generated: tmp.curve2straight.nii.gz
Warping field generated: tmp.straight2curve.nii.gz
Apply transformation to input image…
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -r tmp.centerline_pad_crop.nii.gz -i data.nii -o tmp.anat_rigid_warp.nii.gz -t tmp.curve2straight.nii.gz -n ‘BSpline[3]’ # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314113350.960104-straighten_spinalcord-14jub3zi
Generate output files…
File created: ./warp_curve2straight.nii.gz
File created: ./warp_straight2curve.nii.gz
cp /var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314113350.960104-straighten_spinalcord-14jub3zi/tmp.anat_rigid_warp.nii.gz ./straight_ref.nii.gz
File created: ./data_straight.nii
Remove temporary files…
rm -rf /var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314113350.960104-straighten_spinalcord-14jub3zi
Finished! Elapsed time: 48 s
Done! To view results, type:
fsleyes ./data_straight.nii &
Resample to 0.5mm isotropic…
sct_resample -i data_straight.nii -mm 0.5x0.5x0.5 -x linear -o data_straightr.nii # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314113350.857435-label_vertebrae-ti9i761j
Apply straightening to segmentation…
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i segmentation.nii -r data_straightr.nii -t warp_curve2straight.nii.gz -o segmentation_straight.nii -n Linear # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314113350.857435-label_vertebrae-ti9i761j
sct_maths -i segmentation_straight.nii -thr 0.5 -o segmentation_straight.nii # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314113350.857435-label_vertebrae-ti9i761j
Create label to identify disc…
Creating temporary folder…
Run C2-C3 detector…
C2-C3 detected…
Remove temporary files…
rm -rf /var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314113444.854512-ld0ifbh4
File /var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314113350.857435-label_vertebrae-ti9i761j/labelz.nii.gz already exists. Will overwrite it.
And apply straightening to label…
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i labelz.nii.gz -r data_straightr.nii -t warp_curve2straight.nii.gz -o labelz_straight.nii.gz -n NearestNeighbor # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314113350.857435-label_vertebrae-ti9i761j
Get z and disc values from straight label…
… [255, 3]
Look for template…
Path template: /Users/enriquegomezfigueroa/sct_4.3/data/PAM50
Open template and vertebral levels…
Disc values from template: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19]
Z-values for each disc: [962, 939, 908, 871, 834, 801, 770, 736, 693, 647, 601, 552, 501, 450, 397, 343, 290, 232, 169, 106, 56]
Distances between discs (in voxel): [23.0, 31.0, 37.0, 37.0, 33.0, 31.0, 34.0, 43.0, 46.0, 46.0, 49.0, 51.0, 51.0, 53.0, 54.0, 53.0, 58.0, 63.0, 63.0, 50.0]
Detect intervertebral discs…
Current disc: 2 (z=255). Direction: superior
… Peak found: z=7 (correlation = 0.3446798129799675)
Current disc: 1 (z=293). Direction: superior
… correcting factor: 1.0
… Switching to inferior direction.
Current disc: 3 (z=218). Direction: inferior
… Peak found: z=-4 (correlation = 0.426649759126742)
… correcting factor: 1.1486486486486487
Current disc: 4 (z=172). Direction: inferior
… Peak found: z=-3 (correlation = 0.35060838465960964)
… correcting factor: 1.1711711711711712
Current disc: 5 (z=130). Direction: inferior
… Peak found: z=2 (correlation = 0.3879841966324763)
… correcting factor: 1.1586814086814088
Current disc: 6 (z=96). Direction: inferior
… Peak found: z=6 (correlation = 0.5028242869773111)
… correcting factor: 1.1204935140419012
Current disc: 7 (z=64). Direction: inferior
… Peak found: z=5 (correlation = 0.5461460531736383)
… correcting factor: 1.0955093009172707
Current disc: 8 (z=22). Direction: inferior
… Peak found: z=8 (correlation = 0.532315569367595)
… correcting factor: 1.0685760785270957
Adding top disc based on adjusted template distance: #0
… approximate distance: 25
Un-straighten labeling…
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i segmentation_straight_labeled.nii -r segmentation.nii -t warp_straight2curve.nii.gz -o segmentation_labeled.nii -n NearestNeighbor # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314113350.857435-label_vertebrae-ti9i761j
Clean labeled segmentation (correct interpolation errors)…
File /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314113350.857435-label_vertebrae-ti9i761j/segmentation_labeled.nii already exists. Will overwrite it.
Label discs…
Generate output files…
File created: ./m21_seg_labeled.nii.gz
File created: ./m21_seg_labeled_discs.nii.gz
File created: ./warp_curve2straight.nii.gz
File created: ./warp_straight2curve.nii.gz
File created: ./straight_ref.nii.gz
Remove temporary files…
rm -rf /var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314113350.857435-label_vertebrae-ti9i761j
Done! To view results, type:
fsleyes /Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16/m21.nii.gz -a 100.0 ./m21_seg_labeled.nii.gz -cm subcortical -a 50.0 &
MacBook-QQ:t2_tse_sag_320_16 enriquegomezfigueroa$ sct_register_to_template -i m21.nii.gz -s m21_seg.nii.gz -l m21_seg_labeled.nii.gz -c t2
–
Spinal Cord Toolbox (4.3)
Check template files…
OK: /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_t2.nii.gz
OK: /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_levels.nii.gz
OK: /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_cord.nii.gz
Check parameters:
Data: m21.nii.gz
Landmarks: m21_seg_labeled.nii.gz
Segmentation: m21_seg.nii.gz
Path template: /Users/enriquegomezfigueroa/sct_4.3/data/PAM50
Remove temp files: 1
Check input labels…
ERROR: Label file has 3231 label(s). It must contain one or two labels.
ERROR: Found two labels with same value.
Create temporary folder (/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk)…
Copying input data to tmp folder and convert to nii…
Generate labels from template vertebral labeling
Value = 1 : (70.0, 70.0505921918409, 951.0333382073403) --> ( 70.0, 70.0, 951.0)
Value = 2 : (70.0, 70.30833886308339, 923.9948020349481) --> ( 70.0, 70.0, 924.0)
Value = 3 : (70.0, 70.50121918179356, 889.7335862006682) --> ( 70.0, 71.0, 890.0)
Value = 4 : (70.0, 70.38403779633848, 852.8857673162912) --> ( 70.0, 70.0, 853.0)
Value = 5 : (70.0, 70.30789525112371, 818.2217119405902) --> ( 70.0, 70.0, 818.0)
Value = 6 : (70.0, 70.7014333993707, 786.225032047547) --> ( 70.0, 71.0, 786.0)
Value = 7 : (70.0, 70.80759724209705, 753.9094575256927) --> ( 70.0, 71.0, 754.0)
Value = 8 : (70.0, 70.35343403351428, 715.2977342459287) --> ( 70.0, 70.0, 715.0)
Value = 9 : (70.0, 70.41839798246667, 670.7036147472079) --> ( 70.0, 70.0, 671.0)
Value = 10 : (70.0, 70.41868823000898, 624.8615068668977) --> ( 70.0, 70.0, 625.0)
Value = 11 : (70.0, 70.43996427204287, 577.066096720684) --> ( 70.0, 70.0, 577.0)
Value = 12 : (70.0, 70.4400396432111, 527.0463330029733) --> ( 70.0, 70.0, 527.0)
Value = 13 : (70.0, 70.48122739179244, 475.89622053137083) --> ( 70.0, 70.0, 476.0)
Value = 14 : (70.0, 70.57302038880987, 424.0064011379801) --> ( 70.0, 71.0, 424.0)
Value = 15 : (70.0, 70.64169901804065, 370.32187714089974) --> ( 70.0, 71.0, 370.0)
Value = 16 : (70.0, 70.69621109607577, 316.83818222823635) --> ( 70.0, 71.0, 317.0)
Value = 17 : (70.0, 70.5720703125, 261.3439453125) --> ( 70.0, 71.0, 261.0)
Value = 18 : (70.0, 70.67006390034781, 200.1311979293052) --> ( 70.0, 71.0, 200.0)
Value = 19 : (70.0, 70.47842956120093, 141.44073903002308) --> ( 70.0, 70.0, 141.0)
Value = 20 : (70.0, 70.68440271257172, 92.52008346374544) --> ( 70.0, 71.0, 93.0)
Check if provided labels are available in the template
Binarize segmentation
Done! To view results, type:
fsleyes seg_bin.nii.gz &
Resample data to 1mm isotropic…
load data…
Done! To view results, type:
fsleyes data_1mm.nii &
load data…
Done! To view results, type:
fsleyes seg_bin_1mm.nii.gz &
Position=(149,207,6) – Value= 1
Position=(144,188,6) – Value= 2
Position=(139,161,6) – Value= 3
Position=(139,129,6) – Value= 4
Position=(146,102,5) – Value= 5
Position=(157,79,5) – Value= 6
Position=(170,58,5) – Value= 7
Position=(186,33,5) – Value= 8
Position=(199,10,5) – Value= 9
All labels (useful syntax):
149,207,6,1:144,188,6,2:139,161,6,3:139,129,6,4:146,102,5,5:157,79,5,6:170,58,5,7:186,33,5,8:199,10,5,9
sct_label_utils -i data_1mm.nii -create 102,142,20,1:99,129,20,2:96,111,20,3:96,89,20,4:100,70,17,5:108,54,17,6:117,40,17,7:128,23,17,8:137,7,17,9 -v 1 -o label_projected_1mm.nii.gz # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk
Change orientation of input images to RPI…
Straighten the spinal cord using centerline/segmentation…
Create temporary folder (/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114150.213562-straighten_spinalcord-7rhmvmrd)…
Copy files to tmp folder…
Create the straight space and the safe zone
Length of spinal cord: 234.76689547312802
Size of spinal cord in z direction: 219.0
Ratio length/size: 1.0719949564983013
Safe zone boundaries (curved space): [-157.9805145263672, 61.01948547363281]
Safe zone boundaries (straight space): [-165.86396226293118, 68.9029332101968]
Pad input volume to account for spinal cord length…
Time to generate centerline: 29.0 ms
100%|█████████████████████████████████████████| 236/236 [00:05<00:00, 41.41it/s]
100%|█████████████████████████████████████████| 244/244 [00:10<00:00, 23.61it/s]
Warping field generated: tmp.curve2straight.nii.gz
Warping field generated: tmp.straight2curve.nii.gz
Apply transformation to input image…
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -r tmp.centerline_pad_crop.nii.gz -i data.nii -o tmp.anat_rigid_warp.nii.gz -t tmp.curve2straight.nii.gz -n ‘BSpline[3]’ # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114150.213562-straighten_spinalcord-7rhmvmrd
Generate output files…
File created: ./warp_curve2straight.nii.gz
File created: ./warp_straight2curve.nii.gz
cp /var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114150.213562-straighten_spinalcord-7rhmvmrd/tmp.anat_rigid_warp.nii.gz ./straight_ref.nii.gz
File created: /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/seg_bin_1mm_rpi_crop_straight.nii.gz
Remove temporary files…
rm -rf /var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114150.213562-straighten_spinalcord-7rhmvmrd
Parse list of warping fields…
Check file existence…
OK: /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/data_1mm_rpi.nii
OK: warp_straight2curve.nii.gz
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_ComposeMultiTransform 3 warp_final.nii.gz -R /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/data_1mm_rpi.nii warp_straight2curve.nii.gz # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk
Generate output files…
WARNING: File warp_straight2curve.nii.gz already exists. Deleting it…
File created: warp_straight2curve.nii.gz
Remove unused label on template. Keep only label present in the input label image…
sct_label_utils -i template_label_body.nii.gz -o template_label_body.nii.gz -remove-reference /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/label_projected_1mm_rpi.nii.gz # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk
Dilating input labels using 3vox ball radius
Apply straightening to labels…
Parse list of warping fields…
Get dimensions of data…
43 x 220 x 220 x 1
Apply transformation…
Apply transformation and resample to destination space…
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/label_projected_1mm_rpi_dilate.nii.gz -o /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/label_projected_1mm_rpi_dilate_straight.nii.gz -t warp_curve2straight.nii.gz -r /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/seg_bin_1mm_rpi_crop_straight.nii.gz -n NearestNeighbor # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk
Copy affine matrix from destination space to make sure qform/sform are the same.
Done! To view results, type:
fsleyes /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/seg_bin_1mm_rpi_crop_straight.nii.gz /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/label_projected_1mm_rpi_dilate_straight.nii.gz &
Estimate transformation for step #0…
Labels src: [[-1.107864217544961, 5.8457866761742565, -17.882953550757435], [-3.3371592878478857, 6.338770304640676, -31.555856992120624], [-3.2265677633285534, 6.852030578613281, -50.77251452636719], [-4.241870142573077, 7.1568938879657935, -74.73591020982043], [-1.9794924143141372, 6.236030578613281, -94.98051452636719], [-2.342063518627633, 6.2643324654057295, -111.95221263957473], [-2.3326514959335327, 6.541293736508024, -127.28577768426192], [-2.3589060855539223, 6.771118297911528, -145.77876014040228], [-2.3185997437423396, 6.510678465937218, -162.54389480805733]]
Labels dest: [[-0.0, 46.220001220703125, -86.34002685546875], [-0.0, 46.220001220703125, -99.84002685546875], [-0.0, 45.720001220703125, -116.84002685546875], [-0.0, 46.220001220703125, -135.34002685546875], [-0.0, 46.220001220703125, -152.84002685546875], [-0.0, 45.720001220703125, -168.84002685546875], [-0.0, 45.720001220703125, -184.84002685546875], [-0.0, 46.220001220703125, -204.34002685546875], [-0.0, 46.220001220703125, -226.34002685546875]]
Degrees of freedom (dof): Tx_Ty_Tz_Sz
Optimization terminated successfully.
Current function value: 112.899859
Iterations: 2
Function evaluations: 159
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[ 0. 46.05333455 -152.84002686]
Translation:
[[ -2.58279717 -39.55145289 62.00798296]]
Concatenate transformations: curve --> straight --> affine…
Parse list of warping fields…
Check file existence…
OK: template.nii
OK: warp_curve2straight.nii.gz
OK: straight2templateAffine.txt
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_ComposeMultiTransform 3 warp_final.nii.gz -R template.nii straight2templateAffine.txt warp_curve2straight.nii.gz # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk
Generate output files…
File created: warp_curve2straightAffine.nii.gz
Apply transformation…
Parse list of warping fields…
Get dimensions of data…
43 x 220 x 220 x 1
Apply transformation…
Apply transformation and resample to destination space…
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/data_1mm_rpi.nii -o /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/data_1mm_rpi_straightAffine.nii -t warp_curve2straightAffine.nii.gz -r template.nii -n ‘BSpline[3]’ # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk
Copy affine matrix from destination space to make sure qform/sform are the same.
Done! To view results, type:
fsleyes template.nii /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/data_1mm_rpi_straightAffine.nii &
Parse list of warping fields…
Get dimensions of data…
43 x 220 x 220 x 1
Apply transformation…
Apply transformation and resample to destination space…
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/seg_bin_1mm_rpi_crop.nii.gz -o /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/seg_bin_1mm_rpi_crop_straightAffine.nii.gz -t warp_curve2straightAffine.nii.gz -r template.nii -n Linear # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk
Copy affine matrix from destination space to make sure qform/sform are the same.
Done! To view results, type:
fsleyes template.nii /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/seg_bin_1mm_rpi_crop_straightAffine.nii.gz &
Part II:
Crop data in template space (for faster processing)…
Sub-sample in z-direction (for faster processing)…
sct_resample -i template_crop.nii -o template_crop_sub.nii -f 1x1x0.25 # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk
sct_resample -i template_seg_crop.nii.gz -o template_seg_crop_sub.nii.gz -f 1x1x0.25 # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk
sct_resample -i /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/data_1mm_rpi_straightAffine_crop.nii -o /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/data_1mm_rpi_straightAffine_crop_sub.nii -f 1x1x0.25 # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk
sct_resample -i /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/seg_bin_1mm_rpi_crop_straightAffine_crop.nii.gz -o /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk/seg_bin_1mm_rpi_crop_straightAffine_crop_sub.nii.gz -f 1x1x0.25 # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk
Register straight spinal cord to template…
Create temporary folder (/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114243.804847-register-c7703qd0)…
Copying input data to tmp folder and convert to nii…
–
ESTIMATE TRANSFORMATION FOR STEP #1
Registration parameters:
type … imseg
algo … centermassrot
slicewise … 0
metric … MeanSquares
iter … 10
smooth … 0
laplacian … 0
shrink … 1
gradStep … 0.5
deformation … 1x1x0
init …
poly … 5
filter_size … 5
dof … Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY … 2
rot_method … pcahog
Create temporary folder (/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114244.216976-register-t7d5oadw)…
Copy input data to temp folder…
sct_convert -i src.nii -o /var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114244.216976-register-t7d5oadw/src.nii
sct_convert -i src_seg.nii -o /var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114244.216976-register-t7d5oadw/src_seg.nii
sct_convert -i dest_RPI.nii -o /var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114244.216976-register-t7d5oadw/dest.nii
sct_convert -i dest_seg_RPI.nii -o /var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114244.216976-register-t7d5oadw/dest_seg.nii
Get image dimensions of destination image…
matrix size: 141 x 141 x 80
voxel size: 0.5mm x 0.5mm x 1.99375mm
Split input segmentation…
Split destination segmentation…
Split input image…
Split destination image…
Estimate cord angle for each slice: 0%| | 0/80 [00:00<?, ?iter/s]Switched to method ‘hog’ for slice: 0
Estimate cord angle for each slice: 1%|▎ | 1/80 [00:00<00:42, 1.88iter/s]Switched to method ‘hog’ for slice: 1
Switched to method ‘hog’ for slice: 3
Switched to method ‘hog’ for slice: 4
Estimate cord angle for each slice: 6%|█▋ | 5/80 …
Generate warping field…
–> step1Warp.nii.gz
Generate warping field…
–> step1InverseWarp.nii.gz
Move warping fields…
cp step1Warp.nii.gz /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114243.804847-register-c7703qd0
cp step1InverseWarp.nii.gz /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114243.804847-register-c7703qd0
rm -rf /var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114244.216976-register-t7d5oadw
–
ESTIMATE TRANSFORMATION FOR STEP #2
Apply transformation from previous step
Parse list of warping fields…
Get dimensions of data…
141 x 141 x 80 x 1
Apply transformation…
Apply transformation and resample to destination space…
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i src_seg.nii -o src_seg_reg.nii -t warp_forward_1.nii.gz -r dest_seg_RPI.nii -n NearestNeighbor # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114243.804847-register-c7703qd0
Copy affine matrix from destination space to make sure qform/sform are the same.
Done! To view results, type:
fsleyes dest_seg_RPI.nii src_seg_reg.nii &
Registration parameters:
type … seg
algo … bsplinesyn
slicewise … 0
metric … MeanSquares
iter … 3
smooth … 1
laplacian … 0
shrink … 1
gradStep … 0.5
deformation … 1x1x0
init …
poly … 5
filter_size … 5
dof … Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY … 2
rot_method … pca
sct_image -i dest_seg_RPI.nii -o dest_seg_RPI_pad.nii -pad 0,0,10 # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114243.804847-register-c7703qd0
Estimate transformation
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsRegistration --dimensionality 3 --transform ‘bsplinesyn[0.5,1,3]’ --metric ‘MeanSquares[dest_seg_RPI_pad.nii,src_seg_reg.nii,1,4]’ --convergence 3 --shrink-factors 1 --smoothing-sigmas 1mm --restrict-deformation 1x1x0 --output ‘[step2,src_seg_reg_regStep2.nii]’ --interpolation ‘BSpline[3]’ --verbose 1 # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114243.804847-register-c7703qd0
Concatenate transformations…
Parse list of warping fields…
Check file existence…
OK: dest.nii
OK: warp_forward_1.nii.gz
OK: warp_forward_2.nii.gz
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_ComposeMultiTransform 3 warp_final.nii.gz -R dest.nii warp_forward_2.nii.gz warp_forward_1.nii.gz # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114243.804847-register-c7703qd0
Generate output files…
File created: warp_src2dest.nii.gz
Parse list of warping fields…
Check file existence…
OK: src.nii
OK: warp_inverse_2.nii.gz
OK: warp_inverse_1.nii.gz
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_ComposeMultiTransform 3 warp_final.nii.gz -R src.nii warp_inverse_1.nii.gz warp_inverse_2.nii.gz # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114243.804847-register-c7703qd0
Generate output files…
File created: warp_dest2src.nii.gz
Apply transfo source --> dest…
Parse list of warping fields…
Get dimensions of data…
141 x 141 x 80 x 1
Apply transformation…
Apply transformation and resample to destination space…
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i src.nii -o src_reg.nii -t warp_src2dest.nii.gz -r dest.nii -n Linear # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114243.804847-register-c7703qd0
Copy affine matrix from destination space to make sure qform/sform are the same.
Done! To view results, type:
fsleyes dest.nii src_reg.nii &
Apply transfo dest --> source…
Parse list of warping fields…
Get dimensions of data…
141 x 141 x 80 x 1
Apply transformation…
Apply transformation and resample to destination space…
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i dest.nii -o dest_reg.nii -t warp_dest2src.nii.gz -r src.nii -n Linear # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114243.804847-register-c7703qd0
Copy affine matrix from destination space to make sure qform/sform are the same.
Done! To view results, type:
fsleyes src.nii dest_reg.nii &
Generate output files…
File created: data_1mm_rpi_straightAffine_crop_sub_reg.nii
File created: template_crop_sub_reg.nii
File created: warp_data_1mm_rpi_straightAffine_crop_sub2template_crop_sub.nii.gz
File created: warp_template_crop_sub2data_1mm_rpi_straightAffine_crop_sub.nii.gz
Remove temporary files…
rm -rf /var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114243.804847-register-c7703qd0
Concatenate transformations: anat --> template…
Parse list of warping fields…
Check file existence…
OK: template.nii
OK: warp_curve2straightAffine.nii.gz
OK: warp_data_1mm_rpi_straightAffine_crop_sub2template_crop_sub.nii.gz
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_ComposeMultiTransform 3 warp_final.nii.gz -R template.nii warp_data_1mm_rpi_straightAffine_crop_sub2template_crop_sub.nii.gz warp_curve2straightAffine.nii.gz # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk
Generate output files…
File created: warp_anat2template.nii.gz
Concatenate transformations: template --> anat…
Parse list of warping fields…
Check file existence…
OK: data.nii
OK: warp_template_crop_sub2data_1mm_rpi_straightAffine_crop_sub.nii.gz
OK: straight2templateAffine.txt
OK: warp_straight2curve.nii.gz
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_ComposeMultiTransform 3 warp_final.nii.gz -R data.nii warp_straight2curve.nii.gz -i straight2templateAffine.txt warp_template_crop_sub2data_1mm_rpi_straightAffine_crop_sub.nii.gz # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk
Generate output files…
File created: warp_template2anat.nii.gz
sct_apply_transfo -i template.nii -o template2anat.nii.gz -d data.nii -w warp_template2anat.nii.gz -crop 0 # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk
sct_apply_transfo -i data.nii -o anat2template.nii.gz -d template.nii -w warp_anat2template.nii.gz -crop 0 # in /private/var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk
Generate output files…
File created: warp_template2anat.nii.gz
File created: warp_anat2template.nii.gz
File created: template2anat.nii.gz
File created: anat2template.nii.gz
WARNING: File warp_curve2straight.nii.gz already exists. Deleting it…
File created: warp_curve2straight.nii.gz
WARNING: File warp_straight2curve.nii.gz already exists. Deleting it…
File created: warp_straight2curve.nii.gz
WARNING: File straight_ref.nii.gz already exists. Deleting it…
File created: straight_ref.nii.gz
Delete temporary files…
rm -rf /var/folders/wg/9vmplq4s7llbcbtzm6y4wpsr0000gn/T/sct-20210314114141.330551-register_to_template-be2y71zk
Finished! Elapsed time: 189s
Done! To view results, type:
fsleyes m21.nii.gz template2anat.nii.gz &
Done! To view results, type:
fsleyes /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_t2.nii.gz anat2template.nii.gz &
MacBook-QQ:t2_tse_sag_320_16 enriquegomezfigueroa$ sct_warp_template -d m21.nii.gz -w warp_template2anat.nii.gz -a 0
–
Spinal Cord Toolbox (4.3)
Check parameters:
Working directory … /Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16
Destination image … m21.nii.gz
Warping field … warp_template2anat.nii.gz
Path template … /Users/enriquegomezfigueroa/sct_4.3/data/PAM50
Output folder … label
WARP TEMPLATE:
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_t1.nii.gz -r m21.nii.gz -t warp_template2anat.nii.gz -o label/template/PAM50_t1.nii.gz -n Linear # in /Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_t2.nii.gz -r m21.nii.gz -t warp_template2anat.nii.gz -o label/template/PAM50_t2.nii.gz -n Linear # in /Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_t2s.nii.gz -r m21.nii.gz -t warp_template2anat.nii.gz -o label/template/PAM50_t2s.nii.gz -n Linear # in /Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_cord.nii.gz -r m21.nii.gz -t warp_template2anat.nii.gz -o label/template/PAM50_cord.nii.gz -n NearestNeighbor # in /Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_wm.nii.gz -r m21.nii.gz -t warp_template2anat.nii.gz -o label/template/PAM50_wm.nii.gz -n Linear # in /Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_gm.nii.gz -r m21.nii.gz -t warp_template2anat.nii.gz -o label/template/PAM50_gm.nii.gz -n Linear # in /Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_csf.nii.gz -r m21.nii.gz -t warp_template2anat.nii.gz -o label/template/PAM50_csf.nii.gz -n NearestNeighbor # in /Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_levels.nii.gz -r m21.nii.gz -t warp_template2anat.nii.gz -o label/template/PAM50_levels.nii.gz -n NearestNeighbor # in /Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_levels_continuous.nii.gz -r m21.nii.gz -t warp_template2anat.nii.gz -o label/template/PAM50_levels_continuous.nii.gz -n Linear # in /Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_label_body.nii.gz -r m21.nii.gz -t warp_template2anat.nii.gz -o label/template/PAM50_label_body.nii.gz -n Linear # in /Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_label_disc.nii.gz -r m21.nii.gz -t warp_template2anat.nii.gz -o label/template/PAM50_label_disc.nii.gz -n Linear # in /Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_label_discPosterior.nii.gz -r m21.nii.gz -t warp_template2anat.nii.gz -o label/template/PAM50_label_discPosterior.nii.gz -n Linear # in /Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_spine.nii.gz -r m21.nii.gz -t warp_template2anat.nii.gz -o label/template/PAM50_spine.nii.gz -n Linear # in /Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_centerline.nii.gz -r m21.nii.gz -t warp_template2anat.nii.gz -o label/template/PAM50_centerline.nii.gz -n Linear # in /Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16
/Users/enriquegomezfigueroa/sct_4.3/bin/isct_antsApplyTransforms -d 3 -i /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/PAM50_label_spinal_levels.nii.gz -r m21.nii.gz -t warp_template2anat.nii.gz -o label/template/PAM50_label_spinal_levels.nii.gz -n NearestNeighbor # in /Users/enriquegomezfigueroa/Desktop/Machado Ancona Eugenia/M21/Machado_Ancona_Eugenia_Margarita/Craneo_Cerebro_2017 - 1/t2_tse_sag_320_16
cp /Users/enriquegomezfigueroa/sct_4.3/data/PAM50/template/info_label.txt label/template
Done! To view results, type:
fsleyes m21.nii.gz -cm greyscale -a 100.0 label/template/PAM50_t2.nii.gz -cm greyscale -dr 0 4000 -a 100.0 label/template/PAM50_gm.nii.gz -cm red-yellow -dr 0.4 1 -a 50.0 label/template/PAM50_wm.nii.gz -cm blue-lightblue -dr 0.4 1 -a 50.0 &
hi,
this is what i’ve done:
# segment cord
sct_deepseg_sc -i m21.nii.gz -c t2 -qc qc
# segment vertebral levels
sct_label_vertebrae -i m21.nii.gz -s m21_seg.nii.gz -c t2 -qc qc
# compute CSA per level
sct_process_segmentation -i m21_seg.nii.gz -vert 3:8 -perlevel 1 -vertfile m21_seg_labeled.nii.gz -o csa.csv
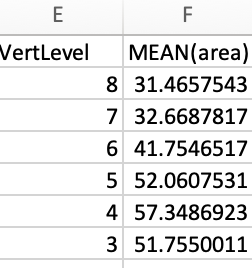
Output CSA:
| Slice (I->S) | VertLevel | MEAN(area) |
|---|---|---|
| 59:79 | 7 | 44.588012478803200 |
| 80:98 | 6 | 49.05501984303600 |
| 99:125 | 5 | 56.86860001785900 |
| 126:146 | 4 | 60.863621026422700 |
| 147:168 | 3 | 47.8760699285787 |
| 169:189 | 2 | 58.94214593820830 |
Output CSV: csa.csv (3.0 KB)
Output QC: qc.zip (886.8 KB)
Two things to note:
- Computing CSA on sagittal images with very thick sliced (3.3mm here) is inaccurate and hence not recommended
- In the QC you will notice slight mis-labeling of the vertebral level. If that happens, you can fix it using the command
sct_label_utilsto label the discs of interest, and then runsct_label_vertebrae -discfile. Example here:
Thank you very much Prof. Cohen,
Just to be clear what I usually do is this:
- sct_propseg -i t2.nii.gz -c t2
- sct_label_vertebrae -i t2.nii.gz -s t2_seg.nii.gz -c t2
- sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -l t2_seg_labeled.nii.gz -c t2
- sct_warp_template -d t2.nii.gz -w warp_template2anat.nii.gz -a 0
- sct_process_segmentation -i t2_seg.nii.gz -vert 3:8 -perlevel 1 -o csa_c3c4.csv
So I should #segment cord (deepest_sc) in step 2?
Hi,
in my previous comment Differences in CSA - #17 by jcohenadad i forgot to add the line with “sct_process_segmentation”. It is now added.
So I should #segment cord (deepest_sc) in step 2?
What I am suggesting is that you bypass registration to template if you don’t need it. And you can do the segmentation of the cord with sct_propseg or sct_deepseg_sc, based on what you find gives the best results.
Again, I’ve updated the code for you in Differences in CSA - #17 by jcohenadad.
It is 3 lines of code.
Run it, let me know if it works for you.