Hi! First, thanks for your wonderful work. SCT works very well!
I have a doubt regarding the segmentations available after template registration to a “subject space”. In short, I have run a pipeline on a bunch of scans consisting of:
- spinal cord segmentation (sct_deepseg)
- labelling of vertebrae (sct_label_vertebrae)
- registration to template (sct_register_to_template)
- warping of template to subject space with previous trasnformation (sct_warp_template)
I was inspecting the segmentations under the obtained template folder, and I don’t quite understand the format of the following file:
template/PAM50_rootlets.nii.gz
The file is not probabilistic (it reaches 8.0), and has continuous values. How should this be interpreted? I would like to obtain a binary (or probabilistic, even better) file with only one class (nerve rootlets). Could anyone tell me how this could be achieved? A threshold of 0.5?
I’ve tried to search for topics covering this issue, but I couldn’t find any. Excuse me if this has already been solved.
Vicent
Dear @vcasellesb,
The sct_warp_template command warps the files found within the PAM50 data folder. So, for background on the template/PAM50_rootlets.nii.gz file, we can look at the changelog for the PAM50 repo: Commits · spinalcordtoolbox/PAM50 · GitHub.
From the list of commits, we can see that the relevant pull request that added this file is Add `PAM50_rootlets.nii.gz` by valosekj · Pull Request #29 · spinalcordtoolbox/PAM50 · GitHub. This pull request contains some background information on the file and how it was generated.
That said, I have also tagged the authors of the changes, and they will be responding shortly with a more detailed reply. 
Kind regards,
Joshua
Dear Joshua,
I appreciate your reply. I’ve seen the PR request you have linked, and the github repo they link there, but I cannot see clear documentation on how the segmentation is encoded.
My guess is that it works similar to template/PAM50_levels_continuous.nii.gz, but there is something that throws me off. In contrast to levels_continuous, in rootlets, within a spinal level (or rootlet) j, the segmentation reaches values below what you would expect (j - 1, j + 1), to values below 1, leading me to believe this is a probabilistic setting.
Also, in template/info_label.txt, it says:
16, spinal dorsal nerve rootlets with discrete spinal levels, PAM50_rootlets.nii.gz
Thanks for tagging the authors, I think their input is needed.
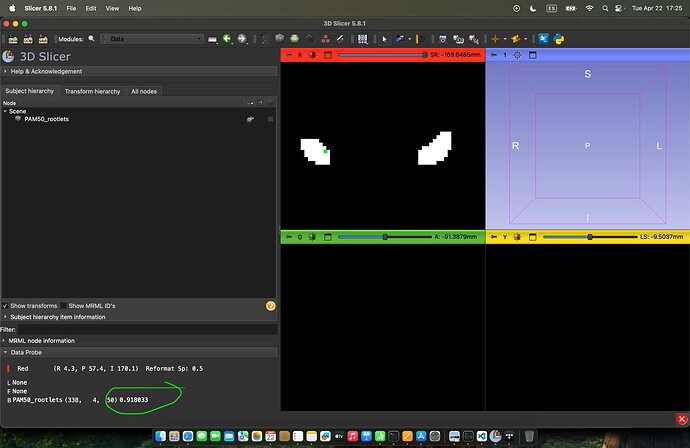
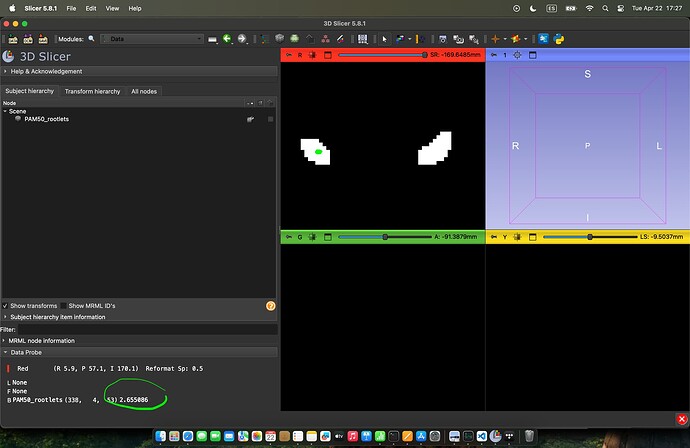
Here I’m showing an example result, from 3D slicer. I’ve marked (axial plane) in green where I’m at with my cursor. Also please see the value that it shows.:
Other values are higher:
Thanks again for your reply. Best,
Vicent
Dear Vicent,
Thank you for providing the details.
The primary purpose of template/PAM50_rootlets.nii.gz is to enable rootlet-based registration to the PAM50 template (recently introduced to SCT in https://github.com/spinalcordtoolbox/spinalcordtoolbox/pull/4647).
What’s your goal in warping PAM50_rootlets.nii.gz from the template to subject space? If you’re interested in spinal levels, you could use PAM50_spinal_levels.nii.gz. Alternatively, you can segment rootlets directly in the subject space.
Let me know more about your use case; happy to help further.
Best,
Jan
Dear Jan,
Thanks so much for your reply. I see, I misunderstood the rootlets file’s purpose.
What I wanted to do is: using the PAM50-derived segmentations of the different regions in the spinal cord (wm, gm, csf, vertebrae) in each native space (and thus with some variability in morphology), to generate a lot of synthetic scans – using a method that uses segmentations as input. I wanted to avoid manual segmentation, honestly.
The objective behind including the rootlets is to have a more precise anatomical parcellation of the spinal cord, and thus obtain more anatomically precise synthetic scans. But honestly, I wanted to map the rootlets to the white matter in the final segmentation (i.e. assign to the nerve rootlets the WM’s value), so that they share the same properties in the synthetic scan. I hope I’ve been clear and made sense.
What would you recommend? I was thinking of applying a threshold, would that make sense? I’ve seen that mostly the rootlets file intersects with the CSF and WM probabilistic maps. The latter is not a problem, since I’m thinking of assigning them to that class anyway. However, the CSF’s case might be a bit more problematic. I’ll probably end up overriding either class, I’m still undecided on which one should take precedence.
Thanks so much for your attention and help. Best regards,
Vicent
Dear Vicent,
Thank you for the additional context. I believe you have already come across SynthSeg for generating synthetic scans from labels? I have tried using SynthSeg for generating spinal cord segmentations in the past, but, my experiments with their default parameters were unsuccessful (see page 7 for a few results).
Assuming you are happy with the segmenations generated by SCT, have you considered simply generating synthetic scans using generative models that don’t require labels? For instance, latent diffusion models (LDM) could be used to generate MRI scans (see this paper using LDMs for brain images). That, of course, depends on the size of the dataset you currently have but you could also consider GANs for image generation. The ultimate idea here is that once you have a dataset of synthetic scans without labels (obtained with GANs or LDMs), you could continue using SCT to generate segmentations on both your real dataset and synthetic dataset.
Happy to discuss further if you have any more questions.
Best,
Naga.