Hello Everyone,
I have computed DTI using this following commands below.
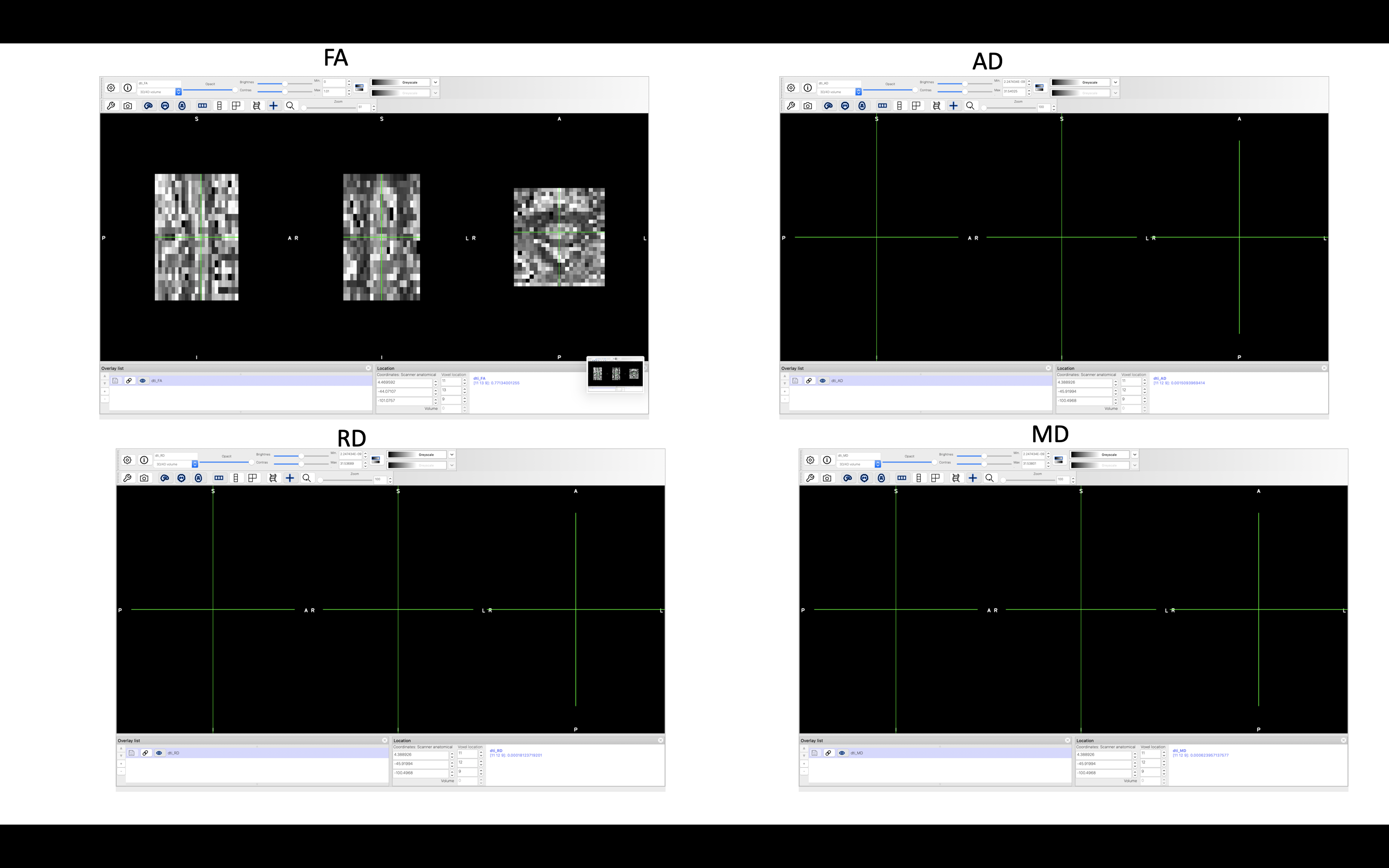
I found that some case did not show MD, RD, and AD on FSL (attached images).
sct_maths -i dmri.nii -mean t -o dmri_mean.nii
sct_propseg -i dmri_mean.nii -c dwi -qc ~/qc_singleSubj
sct_create_mask -i dmri_mean.nii -p centerline,dmri_mean_seg.nii -size 35mm
sct_crop_image -i dmri.nii -m mask_dmri_mean.nii -o dmri_crop.nii
sct_dmri_moco -i dmri_crop.nii -bvec bvecs.txt
sct_propseg -i dmri_crop_moco_dwi_mean.nii -c dwi -qc ~/qc_singleSubj
sct_register_multimodal -i $SCT_DIR/data/PAM50/template/PAM50_t1.nii.gz -iseg $SCT_DIR/data/PAM50/template/PAM50_cord.nii.gz -d dmri_crop_moco_dwi_mean.nii -dseg dmri_crop_moco_dwi_mean_seg.nii -param step=1,type=seg,algo=centermass:step=2,type=seg,algo=bsplinesyn,slicewise=1,iter=3 -initwarp …/t2/warp_template2anat.nii.gz -initwarpinv …/t2/warp_anat2template.nii.gz -qc ~/qc_singleSubj
mv warp_PAM50_t12dmri_crop_moco_dwi_mean.nii.gz warp_template2dmri.nii.gz
mv warp_dmri_crop_moco_dwi_mean2PAM50_t1.nii.gz warp_dmri2template.nii.gz
sct_warp_template -d dmri_crop_moco_dwi_mean.nii -w warp_template2dmri.nii.gz
sct_dmri_compute_dti -i dmri_crop_moco.nii -bval bvals.txt -bvec bvecs.txt
sct_extract_metric -i dti_FA.nii -l 50 -vert 3:4 -o FA_in_sc3_4.csv
Do you know how to fix this?
Thank you again.