Hello.
I need to find dwi metrics in the some slices around the discs (so can’t use neither per-level nor per-slice commends). Is there any way to do so? I am very confused, any help is appreciated.
Thank you
Hi @Roberto_r,
I noticed that you asked a similar question 2 months ago: Upper and lower slices of disc
The example syntax that @jcohenadad had provided in his previous answer to specify slice ranges (sct_process_segmentation -z 165:168) should also work for sct_extract_metric.
Please let me know if you are still confused, and I’ll try to answer. ![]()
Kind regards,
Joshua
Thank you for your response. This can be a solution but there is a problem with that. it cannot accurately estimate the metrics for all images, because the location of discs and their sizes may differ from a patient to another. I am looking for an accurate and automatic solution. For t2, I used below commands and reached the answer. But this method did not work for dwi images, because I do not have the manual labellings of dwi files.
sct_maths -i ${file_t2_seg}_labeled_discs.nii.gz -dilate 3 -o ${file_t2_seg}_labeled_discs_dil.nii.gz
sct_process_segmentation -i ${file_t2_seg}.nii.gz -vert 1:7 -perlevel 1 -vertfile ${file_t2_seg}_labeled_discs_dil.nii.gz -o ${PATH_RESULTS}/csa-SC_T2.csv -append 1
I also tried to automatically extract dwi labellings with the below commands but I received an error.
sct_label_vertebrae -i ${file}.nii.gz -s ${file_seg}.nii.gz -c t1 -discfile ${DISCLABEL}
sct_label_utils -i ${file_seg}_labeled.nii.gz -vert-body 3,5 -o ${FILELABEL}.nii.gz
I am wondering if there is a solution similar to t2 images for extracting the dwi metrics?
Thank you for your time.
Hi @Roberto_r , indeed, for DWI data acquired axially, where discs are not visible, you should instead rely on anatomical images acquired during the same session (assuming minimum subject motion between the anatomical and the DWI scan). So, one possibility is to move the dilated labeled discs you created into the space of the DWI image. This could be done as follows:
sct_register_multimodal -i ${file_t2_seg}_labeled_discs_dil.nii.gz -d DWI_MEAN -identity 1 -x nn
With DWI_MEAN your DWI mean image.
Here we use ‘nn’ because you want your labels to be in integer (not float).
Note that if your slices are very thick, the above command might create “missing” labels in your destination image because of the nearest neighbour interpolation from a fine grid to a coarse grid. If you experience this situation, you can do this:
sct_register_multimodal -i ${file_t2_seg}_labeled_discs_dil.nii.gz -d DWI_MEAN -identity 1 -x nn
sct_apply_transfo -i ${file_t2_seg}_labeled_discs_dil.nii.gz -i DWI_MEAN -w WARP_OUT -x label
With WARP_OUT the output forward warping field of the sct_register_multimodal command
Thanks. I followed the command you suggested, the values that I got e.g. for FA are:
0.044108272679991
0.001616833375825
0.129277906625493
0.039019532181462
0.039968623833716
seems different from standard method to find FA (values around 0.6)
Does this makes sense? should not it still around 0.6 since it’s the average among slices?
Here’s how it looks on dwi:
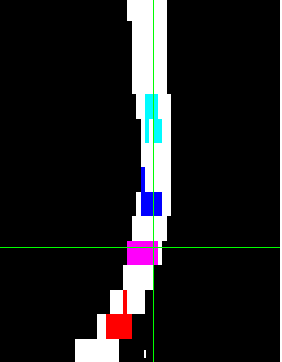
Hi @Roberto_r
It is impossible for me to tell what went wrong in your calculations of FA. It could be the quality of the data, the quality of registration, the syntax used, etc.
For me to be able to help you, please email me your images and the exact code that you used with these images so I can reproduce exactly your results and then I will be able to help
Okay, Thank you. I sent an email to the address you shared ended with polymtl.
Hi Roberto,
Let’s do a step-by-step debugging to understand what went wrong. Firstly, could you please zip and upload your QC folder? That will tell us about the previous steps (segmentation, labeling, etc.)
No follow-up after >5months so I will consider this thread ‘solved’