Hi Julien,
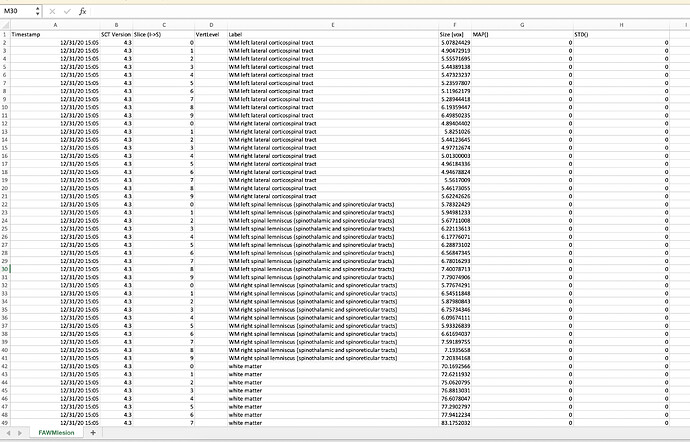
I am analyzing DTI images and I am having trouble with one dataset. I can’t see the FA images on FSLeyes and the FA values are showing 0 on the csv file. I repeated the analysis a couple of times with the same result. Below is the code and some of the screenshots:
Here is my code
Compute mean dMRI from dMRI data
sct_maths -i dmri.nii.gz -mean t -o dmri_mean.nii.gz
Segment SC on mean dMRI data
Note: This segmentation does not need to be accurate-- it is only used to create a mask around the cord
sct_propseg -i dmri_mean.nii.gz -c dwi -qc ~/qc_DTI_Cordotomy
Create mask (for subsequent cropping)
sct_create_mask -i dmri_mean.nii.gz -p centerline,dmri_mean_seg.nii.gz -size 35mm
Crop data for faster processing
sct_crop_image -i dmri.nii.gz -m mask_dmri_mean.nii.gz -o dmri_crop.nii.gz
Motion correction (moco)
sct_dmri_moco -i dmri_crop.nii.gz -bvec bvecs.txt
Segment SC on motion-corrected mean dwi data (check results in the QC report)
sct_propseg -i dmri_crop_moco_dwi_mean.nii.gz -c dwi -qc ~/qc_DTI_Cordotomy
Register template->dwi via t2s to account for GM segmentation
sct_register_multimodal -i $SCT_DIR/data/PAM50/template/PAM50_t1.nii.gz -iseg $SCT_DIR/data/PAM50/template/PAM50_cord.nii.gz -d dmri_crop_moco_dwi_mean.nii.gz -dseg dmri_crop_moco_dwi_mean_seg.nii.gz -param step=1,type=seg,algo=centermass:step=2,type=seg,algo=bsplinesyn,slicewise=1,iter=3 -initwarp warp_template2anat.nii.gz -initwarpinv warp_anat2template.nii.gz -qc ~/qc_DTI_Cordotomy
Rename warping fields for clarity
mv warp_PAM50_t12dmri_crop_moco_dwi_mean.nii.gz warp_template2dmri.nii.gz
mv warp_dmri_crop_moco_dwi_mean2PAM50_t1.nii.gz warp_dmri2template.nii.gz
Warp template
sct_warp_template -d dmri_crop_moco_dwi_mean.nii.gz -w warp_template2dmri.nii.gz -qc ~/qc_DTI_Cordotomy
Check results in the QC report
———————————————————————————
Compute DTI metrics using dipy [1]
sct_dmri_compute_dti -i dmri_crop_moco.nii.gz -bval bvals.txt -bvec bvecs.txt
sct_extract_metric -i dti_FA.nii.gz -z 0:9 -perslice 1 -method map -l 4,5,12,13,51,53,54 -o FAWMlesion.csv