Hello!
I am having errors trying to motion correct fMRI data. I’m looking at func.nii.gz (200 volumes, 256x256x14 slices) collected from Philips 3T MRI, running these commands:
for g in $list; do (
if [ ! -f $g/proc/func_moco.nii.gz ] ; then
sct_fmri_moco -i $g/orig/func.nii.gz -m $g/proc/f_mask30mm.nii.gz
-ofolder $g/proc/ -r 1 -v 2 -x spline
fslmaths $g/proc/func_moco.nii.gz -Tmean $g/proc/func_moco_mean.nii.gz
rm $g/orig/T0
fi
) done
The command runs successfully for the most part, until I receive the following error output:
Loaded datasub_197_mean.nii.gz (/tmp/sct_2025-01-21_20-16-22_moco-wrapper_3cxoji8t/datasub_197_mean.nii.gz) orientation RPI shape (256, 256, 14)
Loaded datasub_198_mean.nii.gz (/tmp/sct_2025-01-21_20-16-22_moco-wrapper_3cxoji8t/datasub_198_mean.nii.gz) orientation RPI shape (256, 256, 14)
Loaded datasub_199_mean.nii.gz (/tmp/sct_2025-01-21_20-16-22_moco-wrapper_3cxoji8t/datasub_199_mean.nii.gz) orientation RPI shape (256, 256, 14)
Image Exception : #63 :: No image files match: 138460/proc/func_moco
terminate called after throwing an instance of ‘std::runtime_error’
what(): No image files match: 138460/proc/func_moco
/cvmfs/neurodesk.ardc.edu.au/containers/fsl_6.0.7.14_20241018/fslmaths: line 3: 82760 Aborted (core dumped) singularity --silent exec --cleanenv --env DISPLAY=$DISPLAY $neurodesk_singularity_opts --pwd “$PWD” /cvmfs/neurodesk.ardc.edu.au/containers/fsl_6.0.7.14_20241018/fsl_6.0.7.14_20241018.simg fslmaths “$@”
rm: cannot remove ‘138460/orig/T0’: No such file or directory
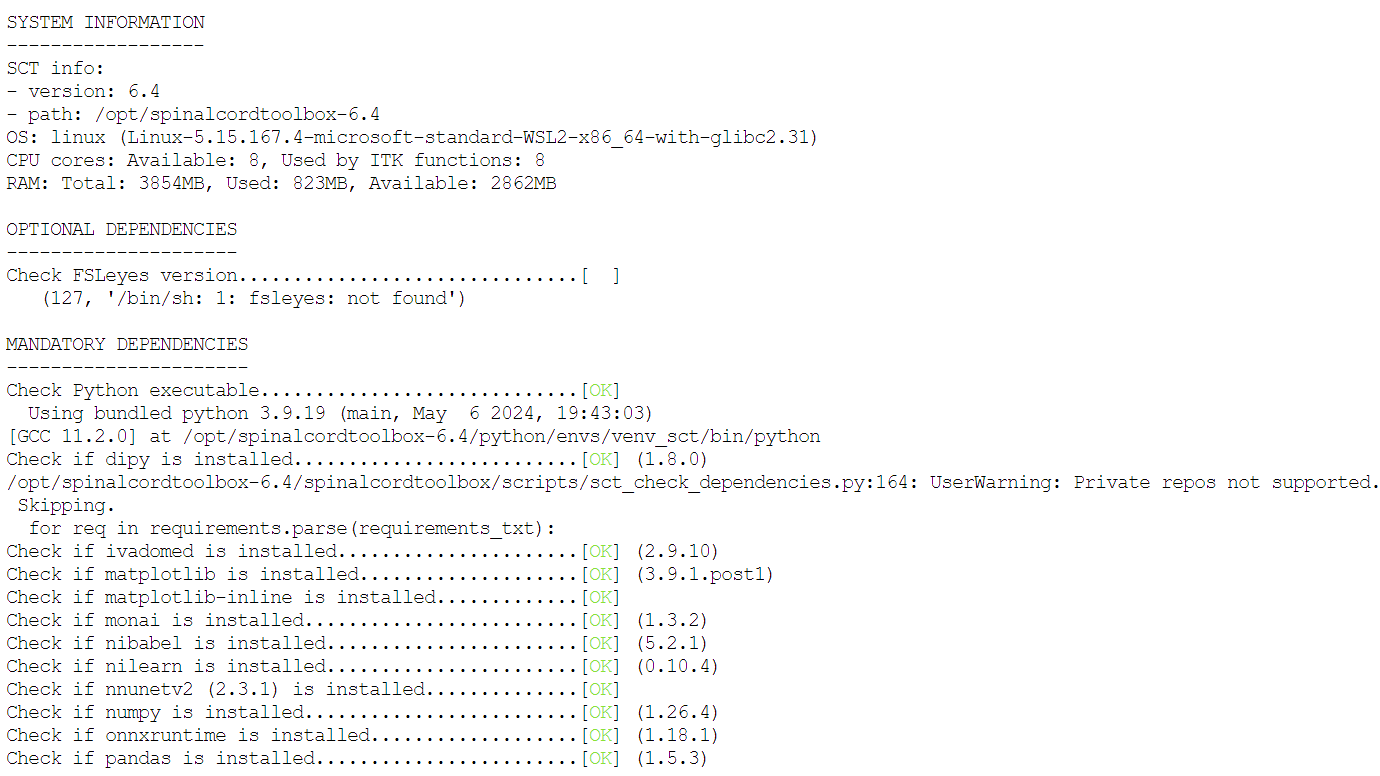
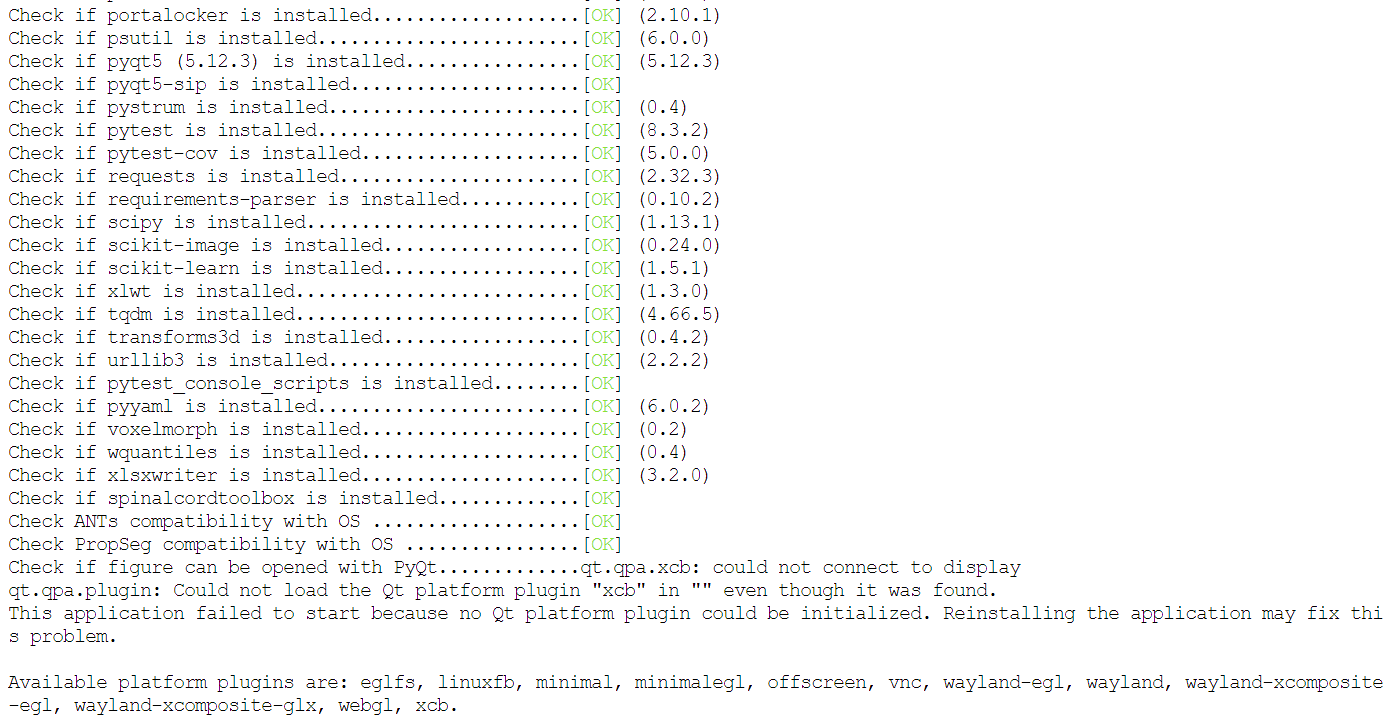
I gathered that the Func_moco file is not being written successfully, but not understanding why. I am running SCT 6.4 and SCT dependencies check showed everything OK except for Qt plugin (screenshot attached). Do you have any recommendations?
Thanks!