Dear SCT experts!
I have found some behaviour I can’t explain to myself, and I need expert advice to figure out what is going on.
Context: we have some relatively poor GRE data (7T) which we want to use for SC segmentation. I have found that sct_deepseg_sc (or propseg) does not find the spinal cord in every slice, but sct_get_centerline is able to find the centerline everyhwere. I don’t understand how one is possible, but not the other.
In depth
I tried segmenting the scans, using T2 contrast, but this segmentation results in some holes, and misses a lot of the thorasic region (not surprising due to the low signal):
sct_deepseg_sc -i GRE.nii -c t2 -qc ./qc -kernel 2d -o seg_2dkernel.nii.gz
I wanted to fill the holes using the -kernel 3d flag, but that made things worse, not better:
sct_deepseg_sc -i GRE.nii -c t2 -qc ./qc -kernel 3d -o seg_3dkernel.nii.gz
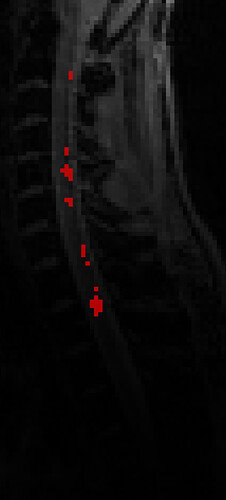
Then I used sct_get_centerline to, well, get the centerline, that worked like a charm*:
Next, I tried using this centerline as an input to help with the segmentation, but it did not help:
sct_deepseg_sc -i GRE.nii -c t2 -file_centerline GRE_centerline_t2.nii.gz -o seg_with_centerline_t2.nii.gz -qc ./qc
Red: segmentation using 2d kernel, without centerline input, yellow: segmentation using 2d kernel with centerline input, white: centerline
The same is observed using 3d kernel:
For completeness sake, I repeated this using T2* contrast. All the data is in the attached zip.
I have a workaround for my problem, so I am asking more for the sake of my understanding. I can see that sct_deepseg_sc is “Finding the spinal cord centerline…”, so I don’t understand what is going on.
Is this just a case of PEBCAC? Is our data this bad?
Best
Daniel
Example_4_SCT.zip (3.0 MB)
Software versions:
OS: Mac OS Monterey 12.3.1
SCT: git-master-3cfab1dbff6a1467ca2fad03d070eb2762dfa5e1