Dear @ryu,
Thank you for the updated data. 
To make sure I am understanding correctly: You are following the GM-informed registration tutorial, right?
In that case, let me summarize the steps needed to complete this tutorial.
- Template registration with the T2 image (Tutorials: Segmentation, Labeling, Registration)
- GM/WM seg of the T2* image (Tutorial)
- GM-informed registration (Tutorial)
I am assuming that you have probably done step 1 and 2 already. But, just to make sure, I will repeat all of the commands that are necessary.
I am assuming that the data is organized like this:
.
├── t2
│ └── t2.nii.gz
└── t2s
└── t2s.nii.gz
Then I would run the following commands:
# 1. Register the T2 data to the template
cd t2
sct_deepseg_sc -i t2.nii.gz -c t2 -qc ~/qc_singleSubj
sct_label_vertebrae -i t2.nii.gz -s t2_seg.nii.gz -c t2 -qc ~/qc_singleSubj
sct_label_utils -i t2_seg_labeled.nii.gz -vert-body 3,9 -o t2_labels_vert.nii.gz
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -l t2_labels_vert.nii.gz -c t2 -qc ~/qc_singleSubj
# 2. Compute GM/WM segmentation of the T2* image
cd ../t2s
sct_deepseg_gm -i t2s.nii.gz -qc ~/qc_singleSubj
sct_deepseg_sc -i t2s.nii.gz -c t2s -qc ~/qc_singleSubj
sct_maths -i t2s_seg.nii.gz -sub t2s_gmseg.nii.gz -o t2s_wmseg.nii.gz
# 3. GM-informed registration
sct_register_multimodal -i "${SCT_DIR}/data/PAM50/template/PAM50_t2s.nii.gz" \
-iseg "${SCT_DIR}/data/PAM50/template/PAM50_wm.nii.gz" \
-d t2s.nii.gz \
-dseg t2s_wmseg.nii.gz \
-initwarp ../t2/warp_template2anat.nii.gz \
-initwarpinv ../t2/warp_anat2template.nii.gz \
-owarp warp_template2t2s.nii.gz \
-owarpinv warp_t2s2template.nii.gz \
-param step=1,type=seg,algo=rigid:step=2,type=seg,metric=CC,algo=bsplinesyn,slicewise=1,iter=3:step=3,type=im,metric=CC,algo=syn,slicewise=1,iter=2 \
-qc ~/qc_singleSubj
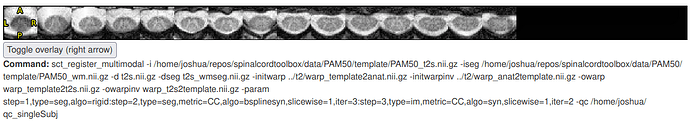
Step 1 looks good. Step 2 looks okay (there is some mis-segmentation but it is not too bad for the purposes of registration). Step 3, however, looks very bad (as you have noticed):
It is not just the QC report, either. Overlaying the images in FSLeyes shows the same large translation.
To fix this, I tried debugging the Step 3 sct_register_multimodal command:
- I edited
-param to remove step=2 and step=3. The large translation still occurs, meaning that it is likely due to step=1.
- I tested the commands against
sct_example_data, and things are okay with our sample data.
- I tested the
-initwarp warping field using sct_warp_template (PAM50_t2s.nii.gz looks OK when warped) and the -initwarpinv warping field on t2s.nii.gz (and that looks OK too)
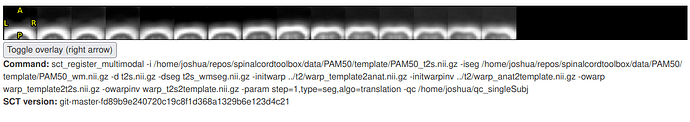
- I changed the
step=1 command to use translation instead of rigid. A large translation still occurs, but in the other direction (strange!):
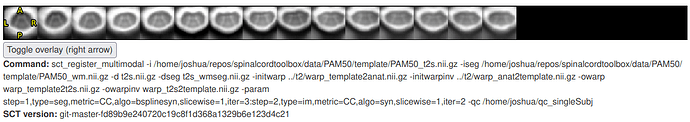
- I tried removing step 1 entirely, and changed the steps
2 -> 1 and 3 -> 2.
So, something is definitely going wrong with the -param step=1,type=seg,algo=rigid part of the command. I will experiment more with the algo options, but I also will invite my colleagues to provide their insight into why rigid and translation may be producing such dramatic translations.
Thank you kindly for your patience and understanding,
Joshua