Description
Hi!
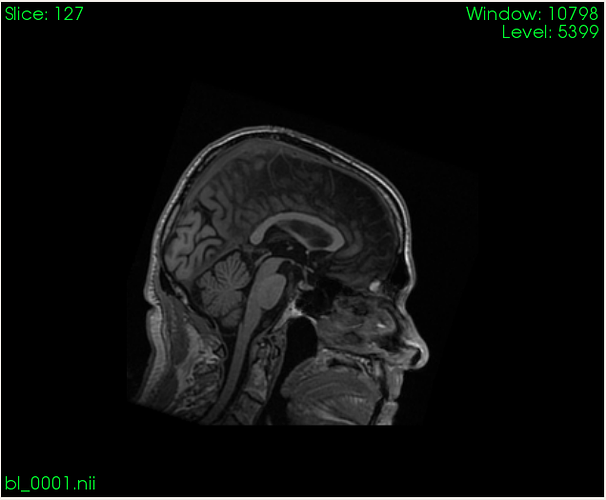
I would like to compute (i.e. extract) volume from specific part of the spinal cord based on a binary mask. Here is what I am working with as an input:
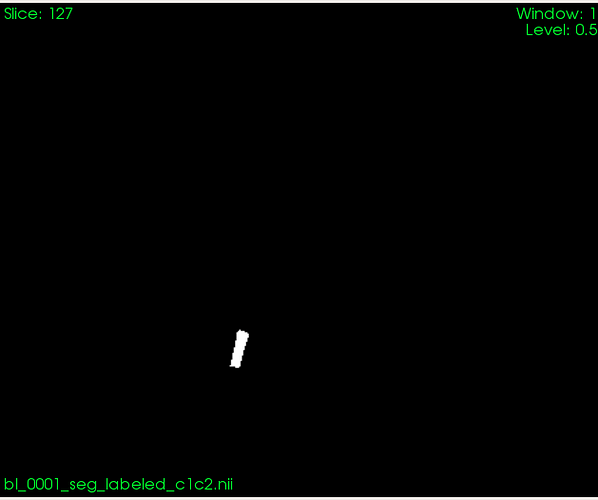
And here is a mask based on which I want to compute volume:
My question is, is this possible to do this with SCT or do I need some external tool (if so, which one)?
Thanks!
Hi @Stefan_Radonjic!
Thank you for reaching out.
What do you mean by “from specific part of the spinal cord”? Did you mean at specific vertebral levels? Eg: your spinal cord segmentation mask would be divided into smaller chunks, and you would compute the volume of each chunk?
The simplest way to compute the volume from your mask is to do the following:
sct_analyze_lesion -m FILE_SEG -s FILE_SEG
Which gives you something like this:
Averaged measures...
Volume [mm^3] = 5780.0+/-0.0
(S-I) Length [mm] = -14.77+/-0.0
Equivalent Diameter [mm] = 6.08+/-0.0
Total volume = 5780.0 mm^3
It also outputs an XLS file.
Also see this related thread: Get spinal cord volume
Cheers
Hi @jcohenadad! Thanks for fast reply!
Well, I think this will work, but to clarify my answer. So, I have 3D T1 image of the brain with portion of the spinal cord. I am able to detect C2-C3 discs on it, but I also label specific portion of the spine myself.
Based on that segmentation, I want to compute volume in the region specified by the segmentation image.
I think that your answer gives me that, just not sure how will this compute voxel volume in the 3D T1 image I am starting with (picture 1 I presented), if I pass segmentation files (-m and -s) for parameters only?
Hi,
I’m sorry but I don’t understand exactly what you are trying to do. For example, would you like to compute the spinal cord volume at the vertebral level C2? (as opposed to in the whole spinal cord segmentation)
Yes, that’s exactly right.
OK, so the easiest way to do it is via the sct_process_segmentation function, which computes the CSA across specified slices, and also the length along the spinal cord axis. So the volume is obtained by multiplying the CSA by the length, across the desired section.
For example, this command:
sct_process_segmentation -i t2_seg.nii.gz -vert 2:5 -vertfile t2_seg_labeled.nii.gz -perlevel 1
Produces this output CSV file:
| Timestamp |
SCT Version |
Filename |
Slice (I->S) |
VertLevel |
MEAN(area) |
STD(area) |
MEAN(angle_AP) |
STD(angle_AP) |
MEAN(angle_RL) |
STD(angle_RL) |
MEAN(diameter_AP) |
STD(diameter_AP) |
MEAN(diameter_RL) |
STD(diameter_RL) |
MEAN(eccentricity) |
STD(eccentricity) |
MEAN(orientation) |
STD(orientation) |
MEAN(solidity) |
STD(solidity) |
SUM(length) |
| 2021-05-08 15:49:49 |
git-master-0e160998510b22bba054b783eb5fb02bad26bfd8 |
/Users/julien/sct_example_data/t2/t2_seg.nii.gz |
123:142 |
5 |
70.4404020395912 |
3.165339525195910 |
1.0998381733640000 |
0.30803310573138400 |
11.159472817536400 |
0.7344994621451710 |
7.262288820889050 |
0.36952222511890100 |
12.485934023396700 |
0.3119704366560290 |
0.8110842295450590 |
0.02956961820477710 |
12.646025854995200 |
2.5255817424078500 |
0.9548061920833870 |
0.014867402258882200 |
16.313040876736400 |
| 2021-05-08 15:49:49 |
git-master-0e160998510b22bba054b783eb5fb02bad26bfd8 |
/Users/julien/sct_example_data/t2/t2_seg.nii.gz |
143:164 |
4 |
75.28273913053960 |
1.831122923241610 |
1.7173171447440500 |
0.057093624422294900 |
11.879833730279900 |
0.3729248045994530 |
7.743264536291100 |
0.2451173130109210 |
12.378087789081900 |
0.26961910178519700 |
0.7787209170044820 |
0.024361284639158600 |
5.411266928681170 |
3.219113990898190 |
0.9644938668858960 |
0.012093599820291500 |
17.993721013739200 |
| 2021-05-08 15:49:49 |
git-master-0e160998510b22bba054b783eb5fb02bad26bfd8 |
/Users/julien/sct_example_data/t2/t2_seg.nii.gz |
165:187 |
3 |
73.20349600691700 |
1.8235438192796300 |
1.2958122048452000 |
0.2921364138287500 |
8.529272398004250 |
1.568547458559680 |
7.9206579168289 |
0.18483796129947200 |
11.769555120942800 |
0.2840328280740710 |
0.738395988126062 |
0.022692478457855400 |
1.1219337088010900 |
0.875112677251581 |
0.9700775123035080 |
0.009623211434786300 |
18.618070488799600 |
| 2021-05-08 15:49:49 |
git-master-0e160998510b22bba054b783eb5fb02bad26bfd8 |
/Users/julien/sct_example_data/t2/t2_seg.nii.gz |
188:206 |
2 |
74.69254399438870 |
2.199616050901140 |
-0.0934919956523575 |
0.49520090153859700 |
4.6772083171808300 |
0.45615364077585600 |
8.213377034419330 |
0.19546718308872300 |
11.525913909952300 |
0.09324897006695110 |
0.7012927666730590 |
0.014861024252918800 |
1.067714651009060 |
1.03883812533707 |
0.9783464998305020 |
0.008655405991361460 |
15.251867063005000 |
Now to get the volume per vertebral level, you can simply multiply column MEAN(area) by column SUM(length), which gives you this (some columns were removed for clarity):
| Slice (I->S) |
VertLevel |
MEAN(area) |
SUM(length) |
Volume (mm^3) |
| 123:142 |
5 |
70.4404020395912 |
16.313040876736400 |
1149.0971578456 |
| 143:164 |
4 |
75.28273913053960 |
17.993721013739200 |
1354.61660506504 |
| 165:187 |
3 |
73.20349600691700 |
18.618070488799600 |
1362.90784868334 |
| 188:206 |
2 |
74.69254399438870 |
15.251867063005000 |
1139.20075160007 |
1 Like
@jcohenadad thank you so much for your help!
1 Like