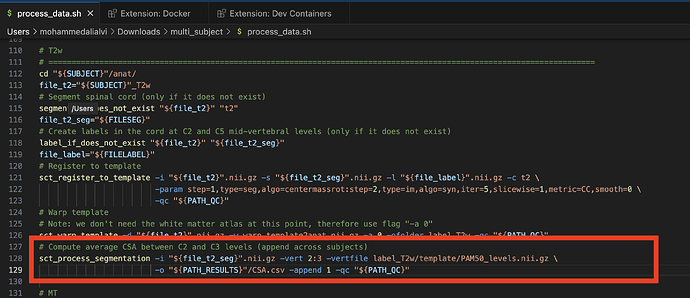
I am trying to process multiple subjects at the same time. I see that in the output, a CSA.csv file is generated which has the CSA area as well as other quantitative metrics for C2-C3 level only. I want to amend the process_data.sh file so that it analyzes ALL levels. Is this possible?
I have attached a screenshot which has the line which computes the C2-C3 CSA. Is there a way this can be amended so that all levels are cervical levels are included ?
Hi @alialvi92,
The part of that command that controls the relevant levels is -vert 2:3. You can change the value of -vert to be, for example, -vert 1:7 instead to cover the cervical levels.
(The numerical vert levels correspond to the levels shown in the figure on this page: Point labeling conventions - Spinal Cord Toolbox documentation)
Also, you can add the option -perlevel 1 and you will get 1 CSA value per level, rather than averaged across all cervical levels. (More details can be found by running sct_process_segmentation -h in your terminal, or checking here.)
Kind regards,
Joshua
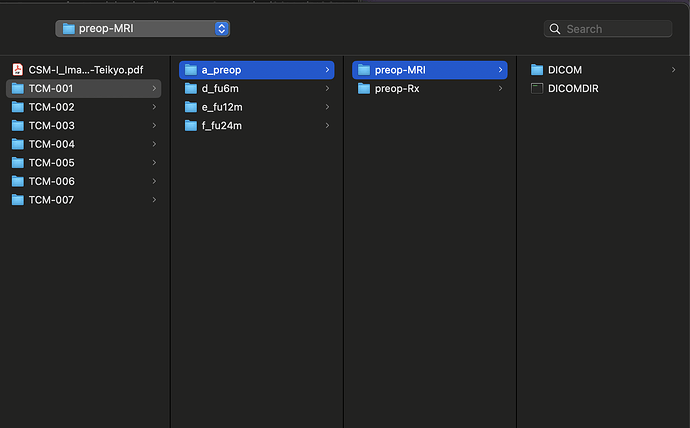
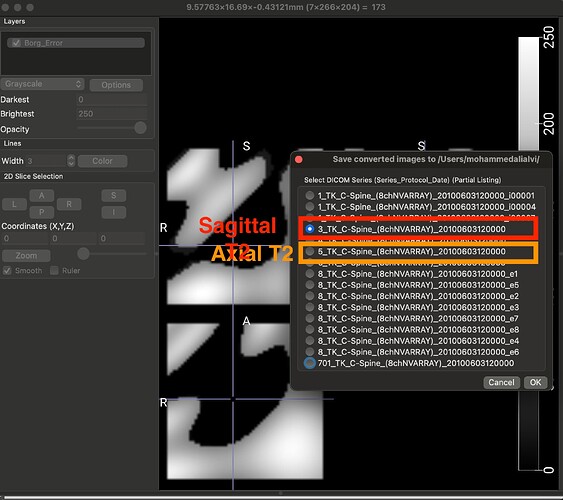
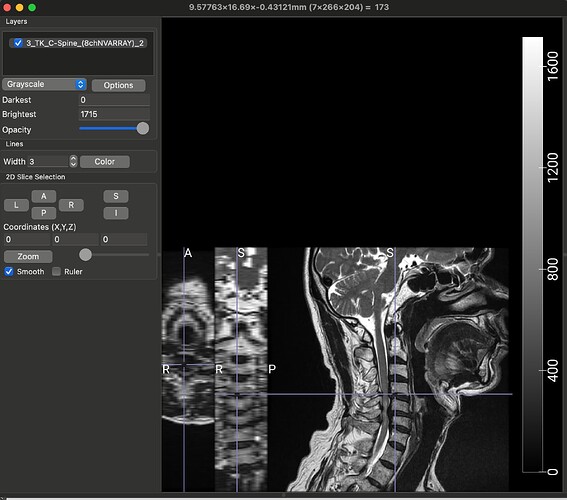
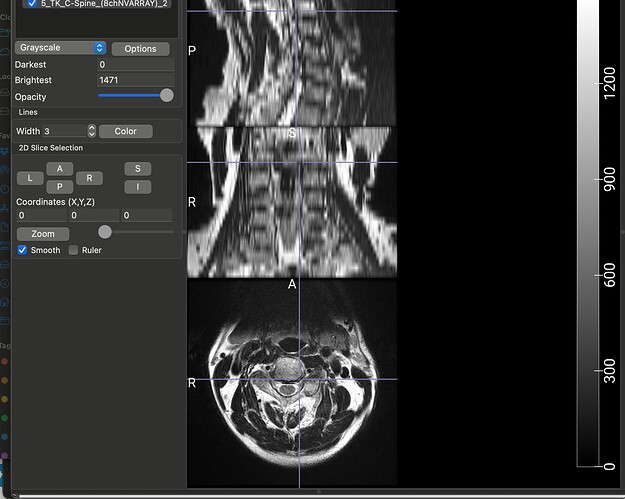
Thank you so much @joshuacwnewton ! That worked perfectly. I have a follow up, though not related. The dataset that I am using for my current analyses, is an older one, hence the spinal MRIs are older (from early 2010s). The files are available in DICOM format (screenshot_1). When I open this file in a DICOM viewer like MRIcroGL, this is what shows up (screenshot_2). Instead of there being a single file representing a T2 sequence, the files are fragmented. So I have highlighted the sagittal T2 sequence and the Axial T2 sequence. When I open these two files, this is how they look (Screenshot_3 and Screenshot_4). My question is: is there a work around here that I can do using sct? There have been instances where the axial T2 sequence is further fragmented into different images and I have used “sct_image -i file_1 file_2 -stitch” to stitch together multiple files in the same plane (axial) into one image. However, I couldn’t find a function to combine sagittal and axial into one? I hope you are able to understand the issue, its rather peculiar I imagine, so sorry if its too confusing.
I have a follow up, though not related
Dear @alialvi92 can you please post your question in another (new) topic, and give a specific title to it, because the title of this current forum thread is not specific to your follow-up question, and will thus be missed by researchers when searching for relevant forum posts. We try to keep this forum organized so that searching topic is more efficient and useful for the community in the long run. Thank you for your understanding.