Hi @willimon,
Thank you very much for your questions!
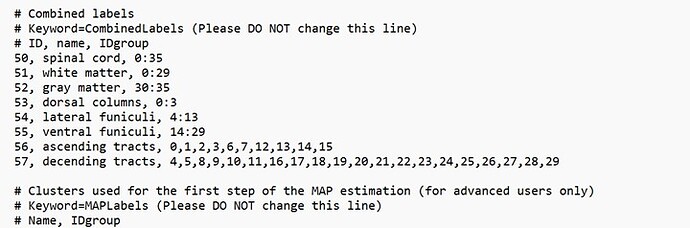
What should I do if I want to add custom tracts, such as ascending tracts,the right and left hemi-cord,etc.? I’ve tried adding custom tracts to the info_lable, as shown in the picture,but that doesn’t work.
Unfortunately, only the “main” ROI files (labels 0:35) are used for sct_analyze_lesion distribution computation. In fact, even the predefined ROIs in “CombinedLabels” (50-55) are not currently considered for sct_analyze_lesion.
I have opened an issue for your feature request:
That being said, I think it would be fairly simple to create a small utility script in Python that “postprocesses” the lesion_analysis.xls file and adds new columns that sum the percentage values corresponding to those within the CombinedLabels. That way, you can calculate the values right now, and don’t have to wait until this feature is added to SCT, nor do you have to fuss with spreadsheet formulas and etc.
If this would be useful for you, I would be happy to write this utility script + provide instructions on how to use it. 
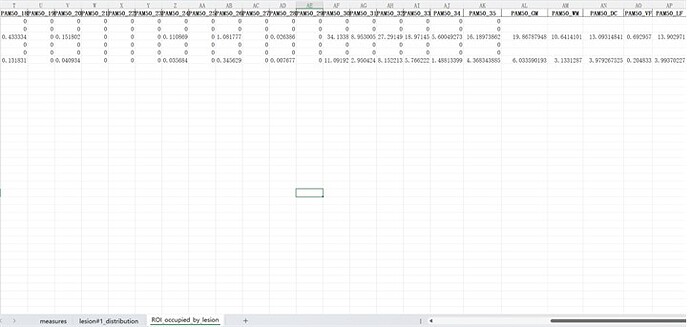
In addition, if I understand correctly, this function is calculated for volume, Is it possible to compute the proportions of each ROI (e.g. GM, WM) occupied by lesions on per axial slice?
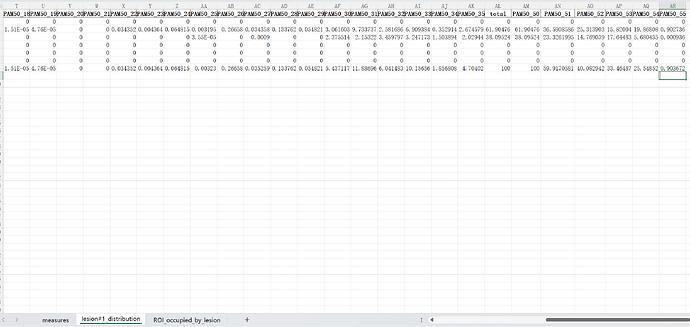
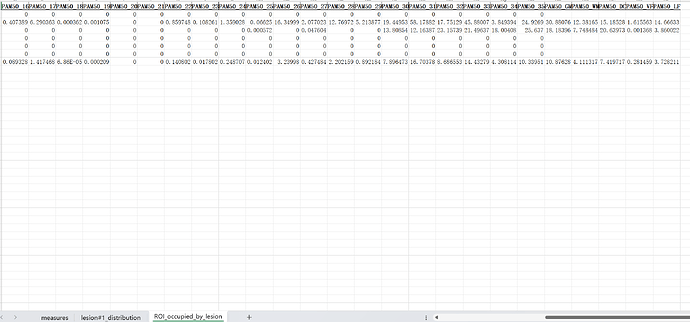
If you look at the lesion_analysis.xls file once more, there is actually a tab called “ROI_occupied_by_lesion” that is very close to what you are requesting.
Unfortunately, rather than being “per axial slice” as you’ve requested, this table is aggregated by vertebral level. So, in order to meet your request, we would need to change the rows to be “slicewise” instead.
As it happens, there is already an open issue related to this feature request:
Given that this previous feature request was also tied to a user’s post on the forum, I will increase the priority of the issue. 
Kind regards,
Joshua