Dear,
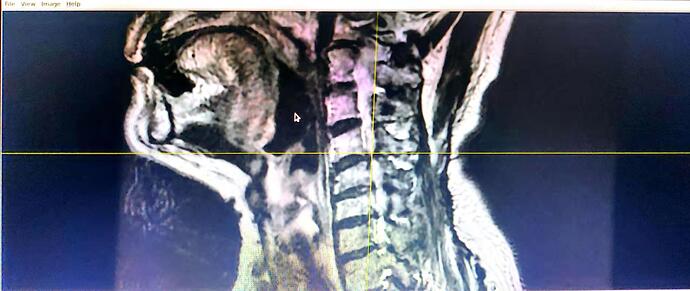
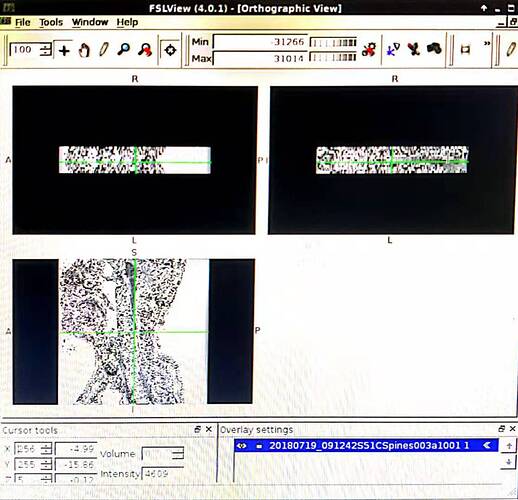
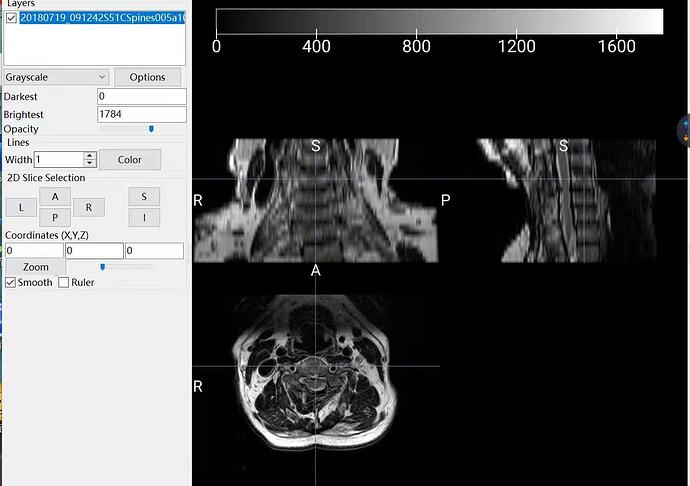
I have a transverse section and coronal position T2* images of one person in 2 folders separately. I use dcm2nii to transfer DICOM to Nifti, however, 2 nii is formed and the pictures of each one are vague. Lower quality compared with the DICOM. The pictures are black and white. Though SCT can be run successfully, limited information can be recognized.
How to transfer DICOM to Nifti and meet the requirement of SCT? Best wishes.
Hi,
It is very difficult for me to understand what the problem could be on your end. If you could share your input DCM data and the resulting NII, it would help us help you.
Cheers,
Julien
Here are the pictures. I appreciate for your help.
Plus. Another problem is that I notice the examples shown in the PDF are 3D. With clear pictures from 3 positions. I try to input all DCM data and hope for output for 1 NII, just like the examples. However, I have not found out how to finish this task.
hum, this is strange. are you able to share the dicom (only for the sagittal scans) so i can try on my end?
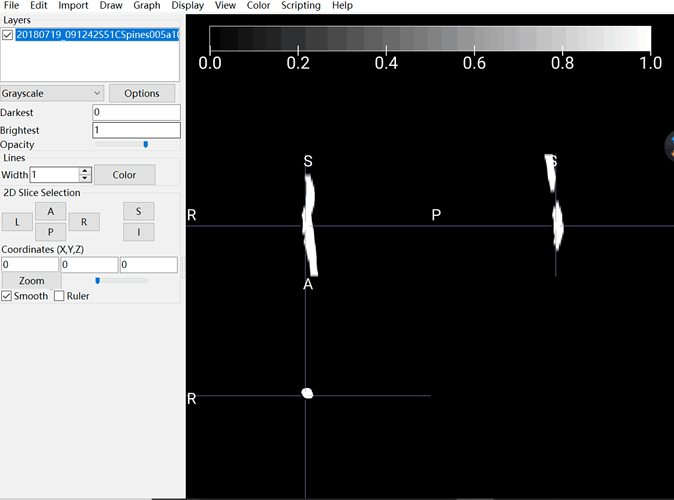
I use dcm2niigui.exe and fix the problem. However, a new problem shows up. There are 23 frames of axial scan. As you can see, the NII pictures do not satisfied.
Hi,
There is only one DICOM file in the link you sent (IMG00001_anon.dcm), so it is expected that the conversion would produce a single 2D nifti sagittal image. But is seems like you do have more dicom files since you have solved your issue using dcm2niigui.exe. If you still experience the problem, please send me all the DICOM files pertaining to a specific sequence (e.g., sagittal multi-slice).
Now, regarding your 2nd problem (the 23 axial frames): what do you mean by “the NII pictures do not satisfied”? What is the criteria you expect to obtain and that is incorrectly converted? If you send me the DICOM files I can try to help.
Best,
Julien
I have a sagittal scan and an axial scan with 4mm slice thickness. After I converted DCM to NII, NII’s resolution is low and SCT output is not well. So I wonder if there is any other solutions or what kind of software you used to convert DCM to NII.
Or what kind of DCM is acceptable? Do you have details about the T2*WI protocols? Such as slice thickness, or do I have to use 3D sequence to obtain DCM?
For example.
Input
Run sct_propseg.
Output
I don’t know why it happens. I suppose it is a result of low quality pictures.
Here is the original NII data.
After I converted DCM to NII, NII’s resolution is low and SCT output is not well. So I wonder if there is any other solutions or what kind of software you used to convert DCM to NII.
I need precise numbers of what is “low” and “high”. What was the resolution on the DCM and on the NII (i.e. voxel size in each dimension)? Please, send me one example of DCM series (not only one sagittal 2D image, but the whole sagittal series). Only sending me the NII is not helpful because I cannot compare the DCM and NII resolution.
Do you have details about the T2*WI protocols?
Here is an example protocol:
do I have to use 3D sequence to obtain DCM?
I’m not sure I understand the question. 2D or 3D acquisition is independent on the output of the scanner. Both will give you a DCM file at the end.
I don’t know why it happens. I suppose it is a result of low quality pictures.
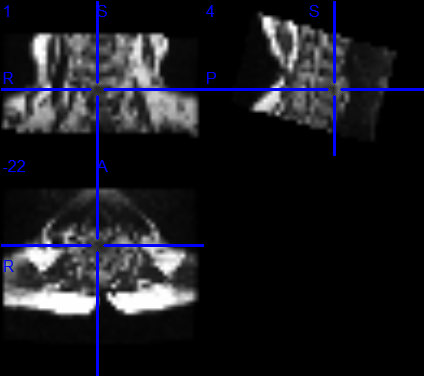
What do you mean by “it”. What is the problem? If you’re talking about the apparent interruption on the coronal view, this is just because you need to navigate across coronal slices. If you are talking about the poor R-L resolution, this is due to your native acquisition resolution (there is nothing I can do about that). One way to mitigate this problem is to resample your image to let’s say 0.8mm isotropic before running propseg.
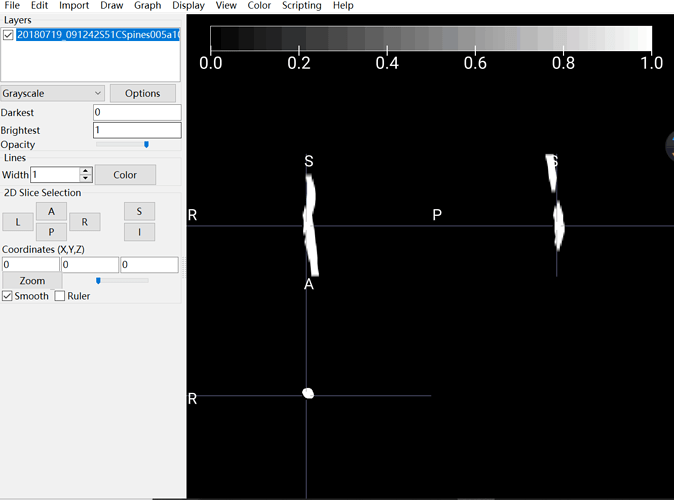
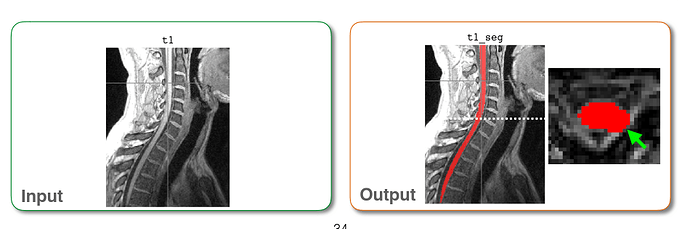
My apology. I didn’t state my question clearly. According to the pdf I download, it is said that after run sct_propseg, input and output are supposed to be colorful, which is totally different from my output:
Input and output mentioned in the PDF for example:
The output I obtained:
As you can see, in my picture, the spinal cord is white and nothing else I can see. I assumed this output should be blamed for my input. That is the reason why I asked about how to convert DCM to NII correctly and accuratly.
Here is the DCM I have. Axial scan and sagittal scan.
The color of the segmentation and the presence of the MRI image in the background depends on the settings of your viewer. It does not depend on the DCM to NII conversion. In the pdf you mentioned, the image was produced using FSLeyes software. For more information about displaying overlay and choosing colors, please see this link.
It seems like you are using another viewer, so I recommend you look at the documentation for the viewer you are using.