Hey Jon,
The most straightforward approach that comes to my mind would be to register your data with the template, and then either (i) use sct_extract_metric with lateral ROIs if you only need to quantify metrics within your mask or (ii) if you need actual masks to be used with your own processing routines, sum up atlas masks corresponding to each lateral ROIs. Example below:
# download example data
sct_download_data -d sct_example_data
sct_example_data/t2s
# segment cord
sct_deepseg_sc -i t2s.nii.gz -c t2s -qc ~/qc
# create label assuming FOV is centered at C3/C4 disc
sct_label_utils -i t2s_seg.nii.gz -create-seg 15,4 -o label_c3c4.nii.gz
# Register to template using input image as reference (we want to avoid straightening due to the thick slices), algo centermassrot to account for rotations, and bsplinesyn algo on the segmentation with iter=5 to allow large and regularized deformations
sct_register_to_template -i t2s.nii.gz -s t2s_seg.nii.gz -ldisc label_c3c4.nii.gz -ref subject -param step=1,type=seg,algo=centermassrot:step=2,type=seg,algo=bsplinesyn,iter=5,slicewise=1,smooth=0 -qc ~/qc
# warp template and atlas
sct_warp_template -d t2s.nii.gz -w warp_template2anat.nii.gz -a 1
At this point, you need to modify the created file label/atlas/info_label.txt to add custom labels corresponding to hemi-cord, by adding the following lines in the # Combined labels section:
56, left hemi-cord, 0,2,4,6,8,10,12,14,16,18,20,22,24,26,28,30,32,34
57, right hemi-cord, 1,3,5,7,9,11,13,15,17,19,21,23,25,27,29,31,33,35
Also see example here: info_label.txt (2.9 KB)
Then, you can use these combined labels to quantify stuff in each hemi-cord:
# quantify metrics on the left side of the cord
sct_extract_metric -i t2s.nii.gz -l 56,57
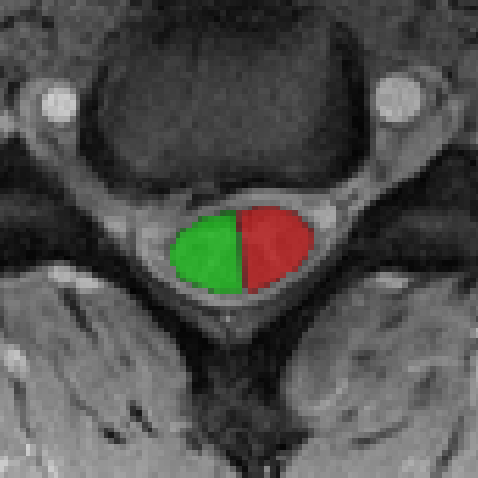
The beauty of this approach versus the R/L section of a segmentation based on the centerline in an FOV coordinate system, is that here the hemi-cord is derived from the registered mask, meaning that the R/L is defined slicewise relative to the cord orientation. Consequently, if the cord is slightly rotated, the output hemi-mask will “truly” be the hemi section of the cord (not a vertical cut which assumes the medial plane of the cord axis is aligned with the sagittal plane of the image).
If you wish to output physical hemi-cord masks (i.e., not use sct_extract_metric), you can generate those as follows:
sct_maths -i label/atlas/PAM50_atlas_00.nii.gz -add $(for i in `seq -w 2 2 34`; do echo "label/atlas/PAM50_atlas_${i}.nii.gz"; done) -o PAM50_atlas_left.nii.gz
sct_maths -i label/atlas/PAM50_atlas_01.nii.gz -add $(for i in `seq -w 3 2 35`; do echo "label/atlas/PAM50_atlas_${i}.nii.gz"; done) -o PAM50_atlas_right.nii.gz
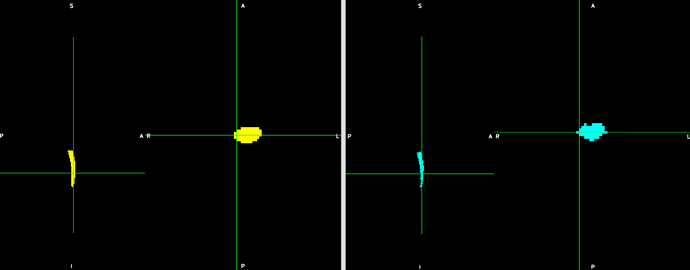
Result looks like this: