Dear SCT team:
I want to extract spinal cord parameters at the level of the intervertebral disc. Do I need to modify my commands. I have previously tried to modify the vertebral body level in some commands to the intervertebral disc level, but the extracted data lacks one intervertebral disc information. I’m not sure if this was caused by wrongly changed commands or the fact that I utilized a sagittal T2 image for template segmentation and registration while using the DTI image for axis registration for later dmri registration. Do these factors affect the normal operation of my program?
Attached are my terminal commands and the final output results.
terminal.txt (182.5 KB)
ad_in_wm.csv (1.0 KB)
fa_in_wm.csv (1.0 KB)
md_in_wm.csv (1.0 KB)
rd_in_wm.csv (1.0 KB)
Note: My deepest apologies for the late response. (The forum’s email notification system has been down, and so we missed your post. The notification system has since been fixed, and we are catching up on missed replies.)
Hi @Shy,
Thank you so much for the question, and for providing the exact commands that you used.
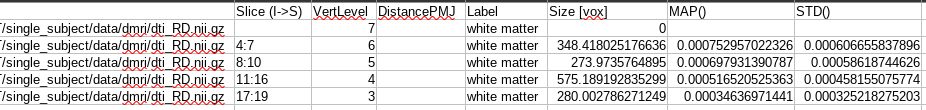
From what I understand, the issue here is that you have specified -vert 3:7 in your extract metric commands, and yet the metric information is missing for vertlevel 7?
The easiest way for us to proceed would be for us to try out each of the commands on your data, so that we can examine the intermediate output between each step.
Are you able to provide each input image file that you used for the initial commands (e.g. t2.nii.gz, dmri.nii.gz, dmri.bvec.txt, etc.)?
Kind regards,
Joshua
Dear SCT team:
It’s okay. During this period, I have reviewed the operation process and introduction on the official website and extracted the values of FA, AD, MD, and RD at the intervertebral disc level. I’m not sure if my code is accurate. If you have time, could you please help me take a look. I will attach my raw data below.
dmri.bval (50 Bytes)
dmri.bvec (226 Bytes)
t2.nii.zip (3.0 MB)
Follows are the commands I used.(Sorry, my dmri.nii.gz file is too large to upload)
sct_deepseg_sc -i t2.nii -c t2 -o t2_seg -qc ~/qc_singleSubj
sct_label_utils -i t2.nii -create-viewer 2,3,4,5,6,7 -o labels_disc.nii.gz \
-msg "Place labels at the posterior tip of each inter-vertebral disc. E.g. Label 2: C2, Label 3: C2/C3, Label 4: C3/C4, Label 5: C4/5, Label 6: C5/6, Label 7: C6/7 etc." (手动标记椎间盘)
sct_label_utils -i labels_disc.nii.gz -vert-body 2,3,4,5,6,7 -o t2_labels_disc.nii.gz
sct_register_to_template -i t2.nii -s t2_seg.nii -l t2_labels_disc.nii.gz -c t2 -qc ~/qc_singleSubj
sct_warp_template -d t2.nii -w warp_template2anat.nii.gz -a 0 -qc ~/qc_singleSubj
sct_process_segmentation -i t2_seg.nii -vert 2:7 -vertfile ./label/template/PAM50_levels.nii.gz -perlevel 1 -o csa_perlevel.csv
sct_dmri_separate_b0_and_dwi -i dmri.nii -bvec dmri.bvec
sct_deepseg_sc -i dmri_dwi_mean.nii -c dwi -qc ~/qc_singleSubj
sct_create_mask -i dmri_dwi_mean.nii -p centerline,dmri_dwi_mean_seg.nii -f cylinder -size 35mm
sct_dmri_moco -i dmri.nii -m mask_dmri_dwi_mean.nii -bvec dmri.bvec \
-qc ~/qc_singleSubj -qc-seg dmri_dwi_mean_seg.nii
sct_deepseg_sc -i dmri_moco_dwi_mean.nii -c dwi -qc ~/qc_singleSub
sct_register_multimodal -i "${SCT_DIR}/data/PAM50/template/PAM50_t1.nii.gz" \
-iseg "${SCT_DIR}/data/PAM50/template/PAM50_cord.nii.gz" \
-d dmri_moco_dwi_mean.nii \
-dseg dmri_moco_dwi_mean_seg.nii \
-initwarp warp_template2anat.nii.gz \
-initwarpinv warp_anat2template.nii.gz \
-owarp warp_template2dmri.nii.gz \
-owarpinv warp_dmri2template.nii.gz \
-param step=1,type=seg,algo=centermass:step=2,type=seg,algo=bsplinesyn,slicewise=1,iter=3 \
-qc ~/qc_singleSubj
sct_warp_template -d dmri_moco_dwi_mean.nii -w warp_template2dmri.nii.gz -qc ~/qc_singleSubj
sct_dmri_compute_dti -i dmri_moco.nii -bval dmri.bval -bvec dmri.bvec
sct_extract_metric -i dti_FA.nii.gz -f label/atlas \
-l 51 -method map \
-vert 2:7 -vertfile label/template/PAM50_levels.nii.gz -perlevel 1 \
-o fa_in_wm.csv
sct_extract_metric -i dti_MD.nii.gz -f label/atlas \
-l 51 -method map \
-vert 2:7 -vertfile label/template/PAM50_levels.nii.gz -perlevel 1 \
-o md_in_wm.csv
sct_extract_metric -i dti_AD.nii.gz -f label/atlas \
-l 51 -method map \
-vert 2:7 -vertfile label/template/PAM50_levels.nii.gz -perlevel 1 \
-o ad_in_wm.csv
sct_extract_metric -i dti_RD.nii.gz -f label/atlas \
-l 51 -method map \
-vert 2:7 -vertfile label/template/PAM50_levels.nii.gz -perlevel 1 \
-o rd_in_wm.csv
Thank you for providing the additional files. ![]()
I believe the dmri.nii.gz is the most important file for debugging your issue, as I can’t perform the motion correction/registration without it. So, if possible, could you upload the dmri.nii.gz file somewhere else? (e.g. Google Drive, Dropbox, etc.)
Kind regards,
Joshua
@Shy Sorry for being late in the game. I understand you want to extract metrics at intervertebral levels (ie: level of the disc), not at the vertebral level (what is currently done with -vert). There is no direct way to do this, but what you could do is input the “disc labels” with -vertfile instead of using the default vertebral labels file.
So, in your commands you listed at How to extract the spinal cord parameters of the intervertebral disc level - #4 by Shy , you should replace:
sct_extract_metric -i dti_FA.nii.gz -f label/atlas \
-l 51 -method map \
-vert 2:7 -vertfile label/template/PAM50_levels.nii.gz -perlevel 1 \
-o fa_in_wm.csv
by:
sct_extract_metric -i dti_FA.nii.gz -f label/atlas \
-l 51 -method map \
-vert 2:7 -vertfile label/template/PAM50_label_disc.nii.gz -perlevel 1 \
-o fa_in_wm.csv
Note however that with the solution I suggested above, you will only get extraction for ONE slice per disc, which is sensitive to noise. In my opinion it would be better to average across 1-2 adjacent slices. @sandrinebedard has written a code to do it.
Note: To run the code, you will need the file
get_disc_slice.py
Hope this is helpful!
Okay, thank you very much for your guidance and suggestions. I will try this new code.
I’m sorry. Yesterday I tried to send these files to you by email, but today it seems that I can’t send emails to your email address. I can’t log on to Google’s relevant website in China, but I only use cloud storage of Baidu online disk. Yesterday, I saw that your colleague had already provided me with relevant suggestions and codes for extracting intervertebral discs. I have decided to give it a try first. If there are any problems, please let me consult with you again. Last, thank you again to you and your team.
Thank you for the concern! No worries at all. I agree, please feel free to try the other suggestion first. ![]()