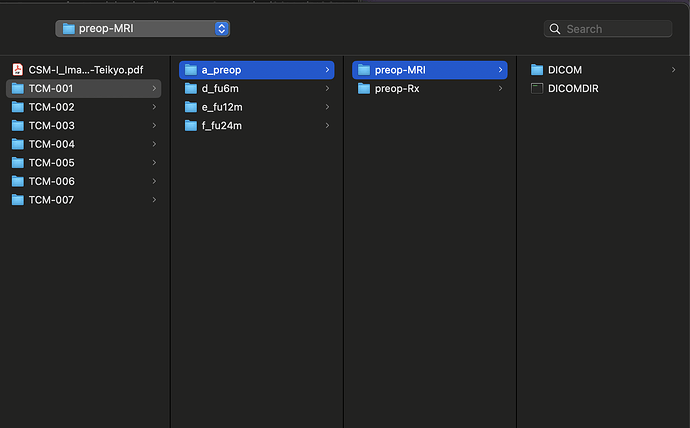
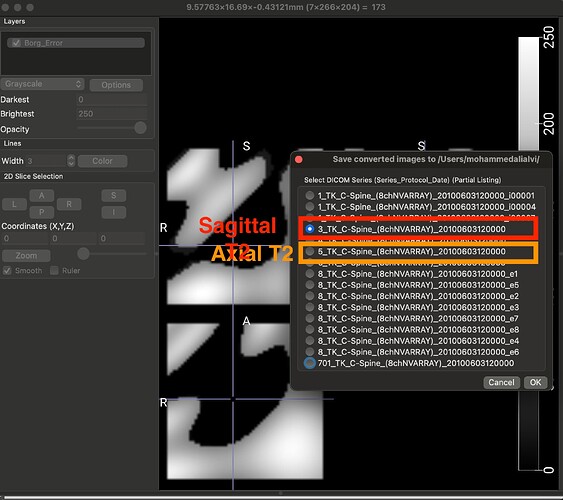
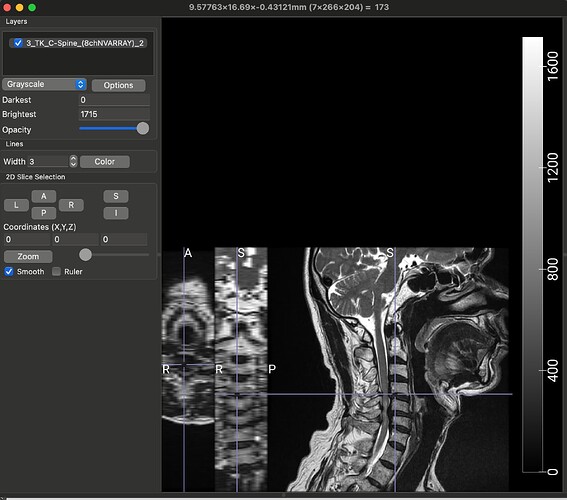
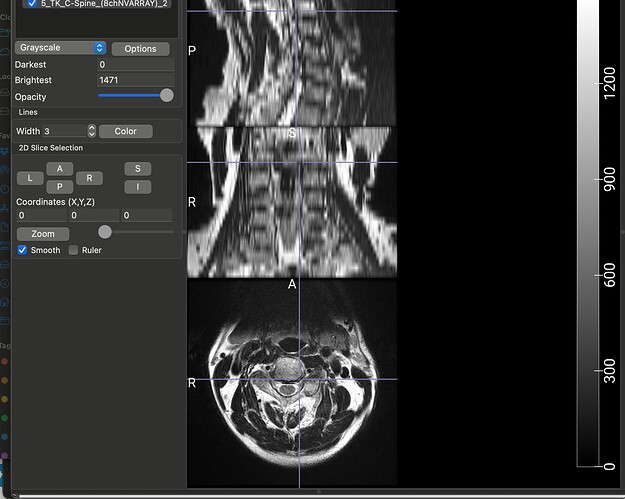
The dataset that I am using for my current analyses, is an older one, hence the spinal MRIs are older (from early 2010s). The files are available in DICOM format (screenshot_1). When I open this file in a DICOM viewer like MRIcroGL, this is what shows up (screenshot_2). Instead of there being a single file representing a T2 sequence, the files are fragmented. So I have highlighted the sagittal T2 sequence and the Axial T2 sequence. When I open these two files, this is how they look (Screenshot_3 and Screenshot_4). My question is: is there a work around here that I can do using sct? There have been instances where the axial T2 sequence is further fragmented into different images and I have used “sct_image -i file_1 file_2 -stitch” to stitch together multiple files in the same plane (axial) into one image. However, I couldn’t find a function to combine sagittal and axial into one? I hope you are able to understand the issue, its rather peculiar I imagine, so sorry if its too confusing.
Hi,
If I understand correctly the issue, you have a flat directory for each patient/session, and inside this directory there are a bunch of DICOM files pertaining to different sequences. And you would like to tease apart each sequence.
For that, you can use dcm2niix.
best
Julien