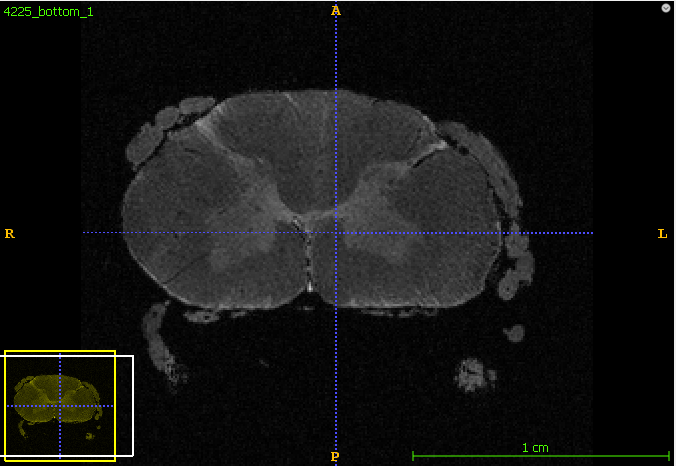
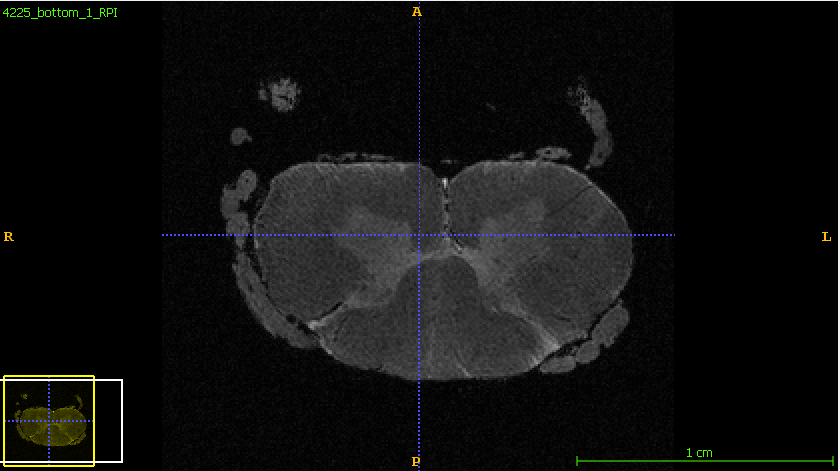
Hi @joshuacwnewton,
Thank you very much for your invaluable information. Thanks again for re-producing the analysis steps. However, I’m still curious why sct_image -setorient RPI not responding while was working in the past (see attached spinal segmentation and header).
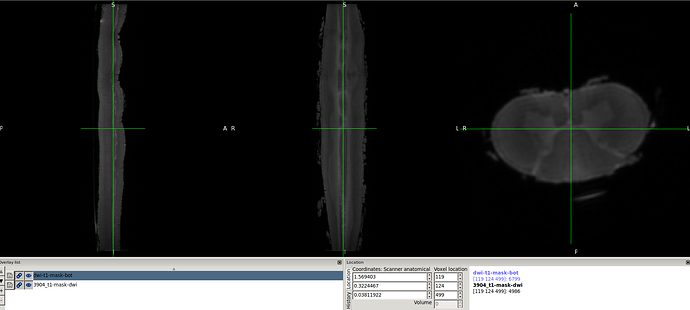
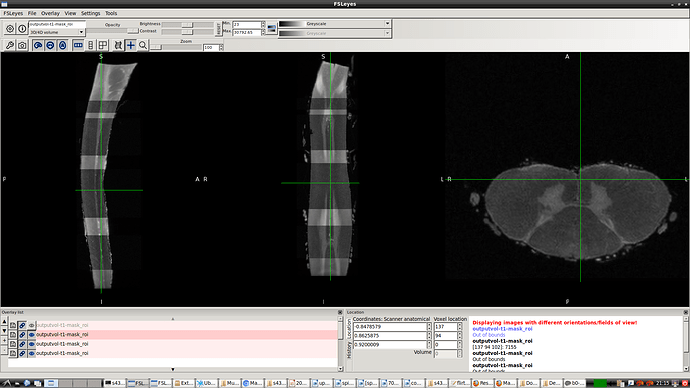
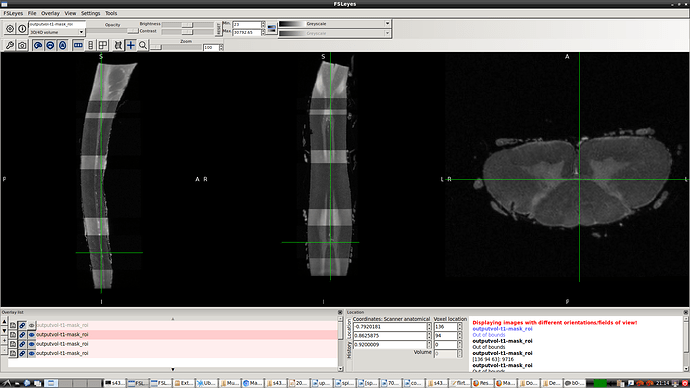
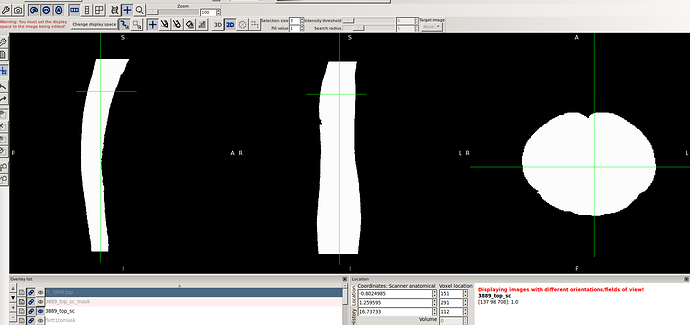
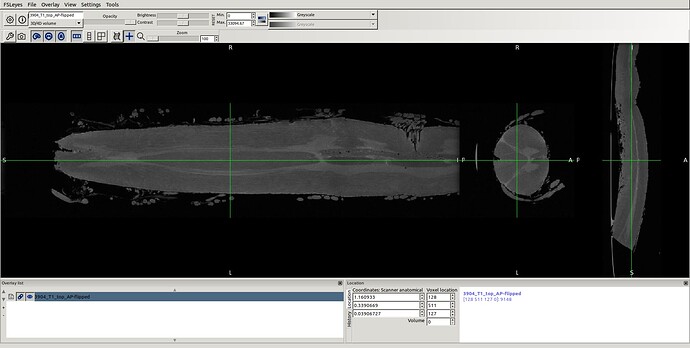
- spinal seg. Header (re-oriented to RPI) that provided by @Charley_Gros which was in ASL orientation before.
sct_image -i 3889_top_sc.nii.gz -header
--
sizeof_hdr 348
data_type FLOAT64
dim [3, 250, 250, 1000, 1, 1, 1, 1]
vox_units mm
time_units s
datatype 64
nbyper 8
bitpix 64
pixdim [-1.0, 0.08, 0.08, 0.08, 1.0, 1.0, 1.0, 1.0]
vox_offset 352
cal_max 0.000000
cal_min 0.000000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
toffset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Scanner Anat
qform_code 1
qto_xyz:1 -0.080000 0.000000 0.000000 10.119999
qto_xyz:2 0.000000 0.080000 0.000000 -6.619999
qto_xyz:3 0.000000 0.000000 0.080000 -39.919998
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Right-to-Left
qform_yorient Posterior-to-Anterior
qform_zorient Inferior-to-Superior
sform_name Scanner Anat
sform_code 1
sto_xyz:1 -0.080000 0.000000 0.000000 10.119999
sto_xyz:2 0.000000 0.080000 0.000000 -6.619999
sto_xyz:3 0.000000 0.000000 0.080000 -39.919998
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Right-to-Left
sform_yorient Posterior-to-Anterior
sform_zorient Inferior-to-Superior
file_type NIFTI-1+
file_code 1
descrip TE=20;sec=36619.0000
aux_file
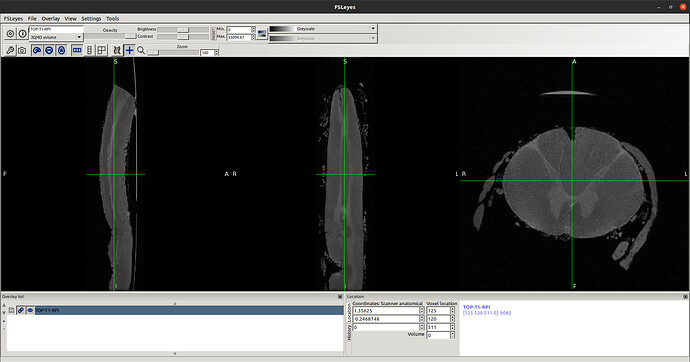
- Raw T1 header before as seen below…
sct_image -i TI_3889.top.nii -header
--
sizeof_hdr 348
data_type UINT16
dim [3, 250, 1000, 250, 1, 1, 1, 1]
vox_units mm
time_units s
datatype 512
nbyper 2
bitpix 16
pixdim [1.0, 0.08, 0.08, 0.08, 0.0, 0.0, 0.0, 0.0]
vox_offset 352
cal_max 0.000000
cal_min 0.000000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
toffset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Scanner Anat
qform_code 1
qto_xyz:1 0.000000 0.000000 0.080000 -9.800000
qto_xyz:2 -0.080000 0.000000 0.000000 13.300000
qto_xyz:3 0.000000 -0.080000 0.000000 40.000000
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Anterior-to-Posterior
qform_yorient Superior-to-Inferior
qform_zorient Left-to-Right
sform_name Scanner Anat
sform_code 1
sto_xyz:1 -0.000000 -0.000000 0.080000 -9.800000
sto_xyz:2 -0.080000 -0.000000 -0.000000 13.300000
sto_xyz:3 0.000000 -0.080000 0.000000 40.000000
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Anterior-to-Posterior
sform_yorient Superior-to-Inferior
sform_zorient Left-to-Right
file_type NIFTI-1+
file_code 1
descrip MRtrix version: 3.0_RC2-7-g4ed6d5ef
aux_file

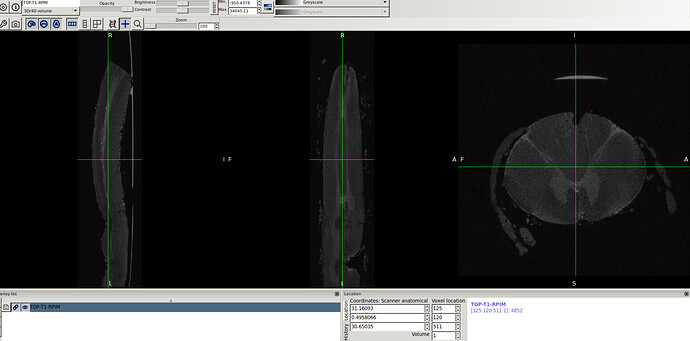
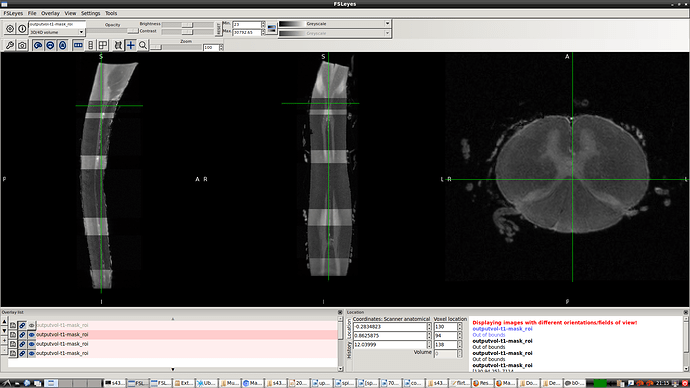
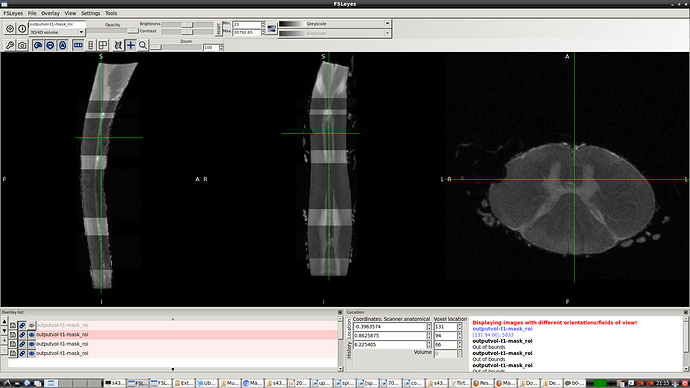
- So, I reproduced same result as you did using sct_image -setorient RPI but no fliping noticed when viewed it. I’ll explain how I got the correct orientation even the method does not seem the correct one. I tried to enforce the data to RPI to test cropping for segment level by co-registering the data to the corrected spinal seg
- I co-registered B0-mean to T1 using the following syntax
flirt -dof 9 -searchrx -180 180 -searchry -180 180 -searchrz -180 180 -in b0-tmean.nii.gz -ref TI_3889.top.nii -omat b0-t1.mat -out flirt-b0-t1.nii.gz
-
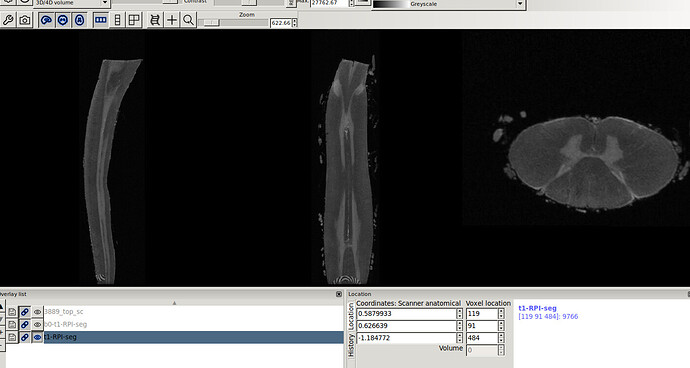
I co-registered the T1 to its RPI seg using
flirt -dof 9 -searchrx -180 180 -searchry -180 180 -searchrz -180 180 -in TI_3889.top.nii -ref 3889_top_sc.nii.gz -omat t1-RPI-seg.mat -out t1-RPI-seg.nii.gz
-
Then, I applied the output transformation t1-RPI-seg.mat to the flirt-b0-t1.nii.gz in step 1 using this syntax
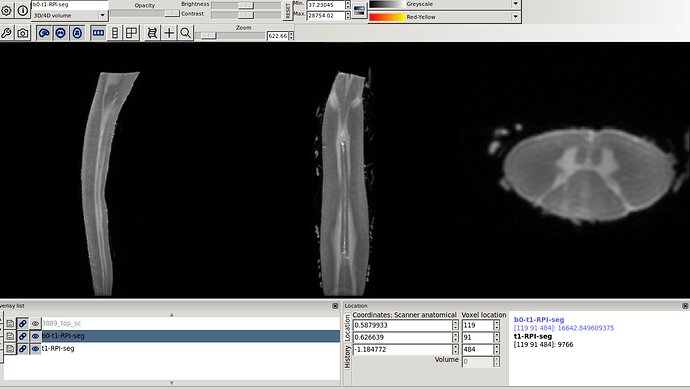
flirt -in flirt-b0-t1.nii.gz -ref 3889_top_sc.nii.gz -applyxfm -init t1-RPI-seg.mat -out b0-t1-RPI-seg.nii.gz
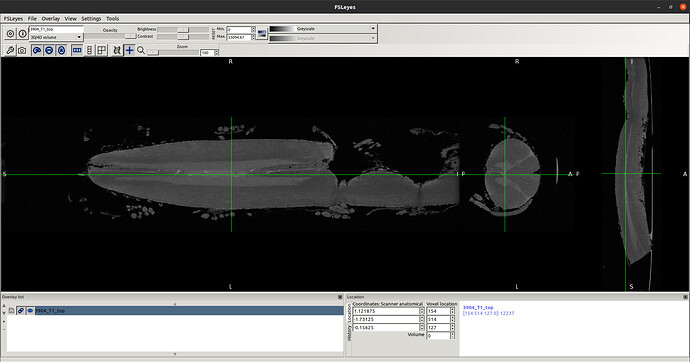
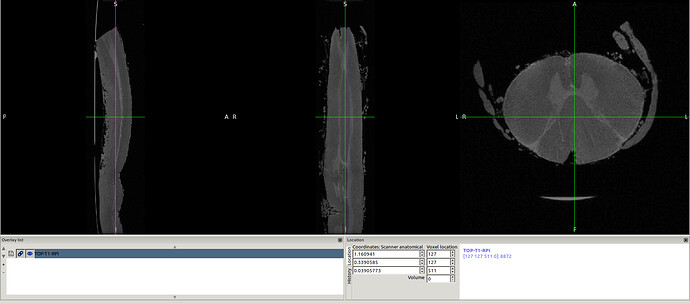
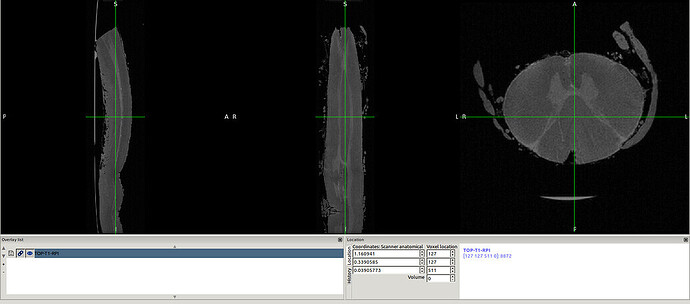
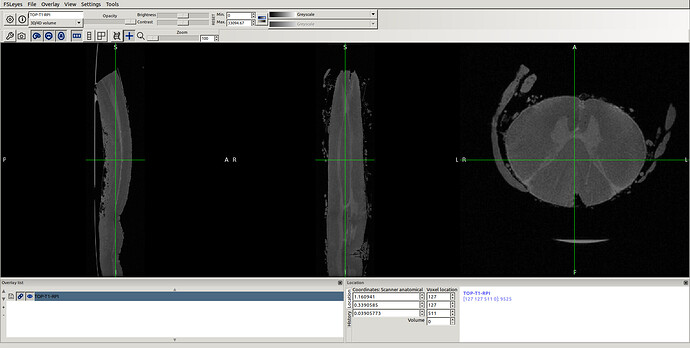
in this way I got the RPI see the new orientation and header info please
- Header for b0-t1-RPI-seg.nii.gz
sct_image -i b0-t1-RPI-seg.nii.gz -header
--
sizeof_hdr 348
data_type FLOAT32
dim [3, 250, 250, 1000, 1, 1, 1, 1]
vox_units mm
time_units s
datatype 16
nbyper 4
bitpix 32
pixdim [-1.0, 0.08, 0.08, 0.08, 0.0, 1.0, 1.0, 1.0]
vox_offset 352
cal_max 0.000000
cal_min 0.000000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
toffset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Scanner Anat
qform_code 1
qto_xyz:1 -0.080000 0.000000 0.000000 10.119999
qto_xyz:2 0.000000 0.080000 0.000000 -6.619999
qto_xyz:3 0.000000 0.000000 0.080000 -39.919998
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Right-to-Left
qform_yorient Posterior-to-Anterior
qform_zorient Inferior-to-Superior
sform_name Scanner Anat
sform_code 1
sto_xyz:1 -0.080000 0.000000 0.000000 10.119999
sto_xyz:2 0.000000 0.080000 0.000000 -6.619999
sto_xyz:3 0.000000 0.000000 0.080000 -39.919998
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Right-to-Left
sform_yorient Posterior-to-Anterior
sform_zorient Inferior-to-Superior
file_type NIFTI-1+
file_code 1
descrip 6.0.5:9e026117
- Header for t1-RPI-seg.nii.gz
sct_image -i t1-RPI-seg.nii.gz -header
--
sizeof_hdr 348
data_type INT32
dim [3, 250, 250, 1000, 1, 1, 1, 1]
vox_units mm
time_units s
datatype 8
nbyper 4
bitpix 32
pixdim [-1.0, 0.08, 0.08, 0.08, 1.0, 1.0, 1.0, 1.0]
vox_offset 352
cal_max 0.000000
cal_min 0.000000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
toffset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Scanner Anat
qform_code 1
qto_xyz:1 -0.080000 0.000000 0.000000 10.119999
qto_xyz:2 0.000000 0.080000 0.000000 -6.619999
qto_xyz:3 0.000000 0.000000 0.080000 -39.919998
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Right-to-Left
qform_yorient Posterior-to-Anterior
qform_zorient Inferior-to-Superior
sform_name Scanner Anat
sform_code 1
sto_xyz:1 -0.080000 0.000000 0.000000 10.119999
sto_xyz:2 0.000000 0.080000 0.000000 -6.619999
sto_xyz:3 0.000000 0.000000 0.080000 -39.919998
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Right-to-Left
sform_yorient Posterior-to-Anterior
sform_zorient Inferior-to-Superior
file_type NIFTI-1+
file_code 1
descrip 6.0.5:9e026117
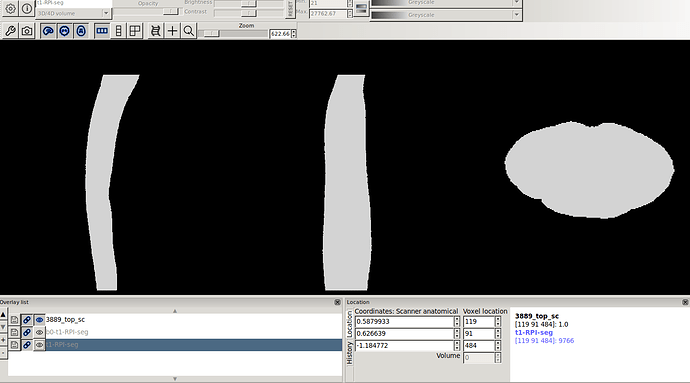
- The above 2 screenshot were re-oriented using the below mask and header.
sct_image -i 3889_top_sc.nii.gz -header
--
sizeof_hdr 348
data_type FLOAT64
dim [3, 250, 250, 1000, 1, 1, 1, 1]
vox_units mm
time_units s
datatype 64
nbyper 8
bitpix 64
pixdim [-1.0, 0.08, 0.08, 0.08, 1.0, 1.0, 1.0, 1.0]
vox_offset 352
cal_max 0.000000
cal_min 0.000000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
toffset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Scanner Anat
qform_code 1
qto_xyz:1 -0.080000 0.000000 0.000000 10.119999
qto_xyz:2 0.000000 0.080000 0.000000 -6.619999
qto_xyz:3 0.000000 0.000000 0.080000 -39.919998
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Right-to-Left
qform_yorient Posterior-to-Anterior
qform_zorient Inferior-to-Superior
sform_name Scanner Anat
sform_code 1
sto_xyz:1 -0.080000 0.000000 0.000000 10.119999
sto_xyz:2 0.000000 0.080000 0.000000 -6.619999
sto_xyz:3 0.000000 0.000000 0.080000 -39.919998
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Right-to-Left
sform_yorient Posterior-to-Anterior
sform_zorient Inferior-to-Superior
file_type NIFTI-1+
file_code 1
descrip TE=20;sec=36619.0000
aux_file
- However, this method has failed in most of my data. Also, I tried the following syntax to copy the desired header to the data that were failed. Unfortunately, it didn’t work out.
“fslcpgeom - copy certain parts of the header information (image dimensions, voxel dimensions, voxel dimensions units string, image orientation/origin or qform/sform info) from one image to another. Note that only copies from Analyze to Analyze or Nifti to Nifti will work properly. Copying from different files will result in loss of information or potentially incorrect settings.”
Could you please review my steps and advice if possible with a solution to this matter?
- BTW, for this advice " then you will first need to reorient the data array only for the input image"
how to achieve it please?
Many thanks in advance.
Best,
Ibrahim