Same error on 2 different scans, when running the function sct_label_vertebrae on a preprocesssed a T2 weighted image and the spinalcord segmentation (created with sct_deepseg_sc),
sct_label_vertebrae -i /tmp/t/preprocessed.nii.gz -s /tmp/spinalcord.nii.gz -c t2 -r 0 -ofolder /tmp/ -qc /tmp/qc_labelvertebrae -v 2
the function fails with this error:
Apply transformation and resample to destination space...
/icometrix-install/spinalcordtoolbox-5.3.0/bin/isct_antsApplyTransforms -d 3 -i segmentation.nii -o segmentation_straight.nii -t warp_curve2straight.nii.gz -r data_straightr.nii -n Linear # in /tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o
Copy affine matrix from destination space to make sure qform/sform are the same.
Loaded segmentation_straight.nii (/tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/segmentation_straight.nii) orientation RPI shape (115, 141, 624)
Loaded data_straightr.nii (/tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/data_straightr.nii) orientation RPI shape (115, 141, 624)
Saving image to /tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/segmentation_straight.nii orientation RPI shape (115, 141, 624)
Loaded segmentation_straight.nii (/tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/segmentation_straight.nii) orientation RPI shape (115, 141, 624)
File /tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/segmentation_straight.nii already exists. Will overwrite it.
Saving image to /tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/segmentation_straight.nii orientation RPI shape (115, 141, 624)
Create label to identify disc...
Loaded data.nii (/tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/data.nii) orientation RPI shape (115, 620, 620)
Loaded segmentation.nii (/tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/segmentation.nii) orientation RPI shape (115, 620, 620)
Smoothing factor: smooth=3.3333333333333335
Smoothing factor: smooth=3.3333333333333335
Smoothing factor: smooth=3.3333333333333335
Smoothing factor: smooth=3.3333333333333335
Creating temporary folder...
Creating temporary folder (/tmp/sct-20220128145619.154167-plptose_)
Saving image to data_midSlice.nii (/tmp/sct-20220128145619.154167-plptose_/data_midSlice.nii) orientation PIR shape (620, 620)
Run C2-C3 detector...
Saving image to pred_midSlice_before_postPro.nii.gz (/tmp/sct-20220128145619.154167-plptose_/pred_midSlice_before_postPro.nii.gz) orientation PIR shape (620, 620)
Saving image to mask_midSlice.nii.gz (/tmp/sct-20220128145619.154167-plptose_/mask_midSlice.nii.gz) orientation PIR shape (620, 620)
Saving image to pred_midSlice_after_postPro.nii.gz (/tmp/sct-20220128145619.154167-plptose_/pred_midSlice_after_postPro.nii.gz) orientation PIR shape (620, 620)
C2-C3 detected...
Temporary files saved to /tmp/sct-20220128145619.154167-plptose_
Saving image to /tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/labelz.nii.gz orientation RPI shape (115, 620, 620)
Loaded /tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/labelz.nii.gz orientation RPI shape (115, 620, 620)
File /tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/labelz.nii.gz already exists. Will overwrite it.
Saving image to /tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/labelz.nii.gz orientation RPI shape (115, 620, 620)
And apply straightening to label...
Parse list of warping fields...
Loaded warp_curve2straight.nii.gz (/tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/warp_curve2straight.nii.gz) orientation RPI shape (115, 141, 624, 1, 3)
Get dimensions of data...
Loaded labelz.nii.gz (/tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/labelz.nii.gz) orientation RPI shape (115, 620, 620)
115 x 620 x 620 x 1
Apply transformation...
Apply transformation and resample to destination space...
/icometrix-install/spinalcordtoolbox-5.3.0/bin/isct_antsApplyTransforms -d 3 -i labelz.nii.gz -o labelz_straight.nii.gz -t warp_curve2straight.nii.gz -r data_straightr.nii -n NearestNeighbor # in /tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o
Copy affine matrix from destination space to make sure qform/sform are the same.
Loaded labelz_straight.nii.gz (/tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/labelz_straight.nii.gz) orientation RPI shape (115, 141, 624)
Loaded data_straightr.nii (/tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/data_straightr.nii) orientation RPI shape (115, 141, 624)
Saving image to /tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/labelz_straight.nii.gz orientation RPI shape (115, 141, 624)
Get z and disc values from straight label...
Loaded labelz_straight.nii.gz (/tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/labelz_straight.nii.gz) orientation RPI shape (115, 141, 624)
.. [530, 0]
Look for template...
Path template: /icometrix-install/spinalcordtoolbox-5.3.0/data/PAM50
Open template and vertebral levels...
Loaded /icometrix-install/spinalcordtoolbox-5.3.0/data/PAM50/template/PAM50_t2.nii.gz orientation RPI shape (141, 141, 991)
Loaded /icometrix-install/spinalcordtoolbox-5.3.0/data/PAM50/template/PAM50_levels.nii.gz orientation RPI shape (141, 141, 991)
Loaded data_straightr.nii (/tmp/sct-20220128144857.530973-label_vertebrae-t2yf2k9o/data_straightr.nii) orientation RPI shape (115, 141, 624)
Disc values from template: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19]
Z-values for each disc: [963, 938, 907, 870, 833, 800, 769, 735, 692, 646, 600, 551, 500, 449, 396, 342, 289, 231, 168, 104, 79]
Distances between discs (in voxel): [25.0, 31.0, 37.0, 37.0, 33.0, 31.0, 34.0, 43.0, 46.0, 46.0, 49.0, 51.0, 51.0, 53.0, 54.0, 53.0, 58.0, 63.0, 64.0, 25.0]
Detect intervertebral discs...
Current disc: -1 (z=530). Direction: superior
.. Peak found: z=0 (correlation = 0.21831008732141663)
Traceback (most recent call last):
File "/icometrix-install/spinalcordtoolbox-5.3.0/spinalcordtoolbox/vertebrae/core.py", line 206, in vertebral_detection
approx_distance_to_next_disc = list_distance[list_disc_value_template.index(current_disc - 1)]
ValueError: -2 is not in list
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/icometrix-install/spinalcordtoolbox-5.3.0/spinalcordtoolbox/scripts/sct_label_vertebrae.py", line 482, in <module>
main(sys.argv[1:])
File "/icometrix-install/spinalcordtoolbox-5.3.0/spinalcordtoolbox/scripts/sct_label_vertebrae.py", line 419, in main
verbose=verbose, path_template=path_template, path_output=path_output, scale_dist=scale_dist)
File "/icometrix-install/spinalcordtoolbox-5.3.0/spinalcordtoolbox/vertebrae/core.py", line 208, in vertebral_detection
logger.warning('Disc value not included in template. Using previously-calculated distance: %s', approx_distance_to_next_disc)
UnboundLocalError: local variable 'approx_distance_to_next_disc' referenced before assignment
Out[8]: 1
This last error should be avoided and a more meaningful error can be printed when there is a value error?
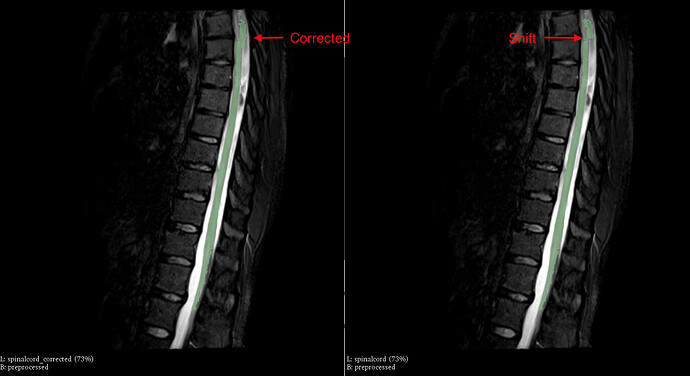
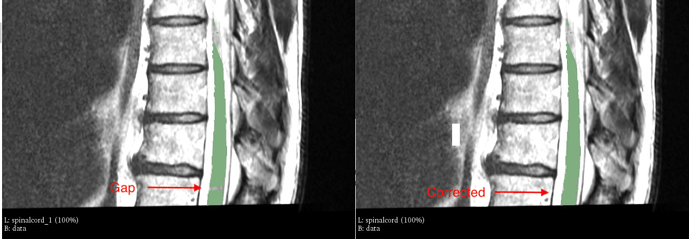
I noticed the spinalcord segmentations are suboptimal (outputs of sct_deepseg_sc).
On 1 example there is a shift on the top part of the spinalcord. In the other example there is a gap in the bottom part of the spinalcord.


Solution:
By manually improving the spinalcord.nii.gz the problem is solved. And the vertebrae is labeled (though incorrect cause no mask/initialization was provided for these thoracic scans)