Hi SCT Team,
When using the sct_label_vertebrae function to automatically identify spinal cord segments, I have observed that slices at the boundary between two segments are sometimes labeled with two values. For instance, at the boundary between C2 and C3, the slice at z=70 is labeled with both values 2 and 3.
I understand that this dual-labeling might be a result of probabilistic factors. However, I am concerned about its impact on subsequent analyses, particularly those involving segmental shape metrics such as CSA.
My question is: Should I remove these boundary slices using custom python codes to prevent potential distortions in segment-specific shape metrics, or is there a recommended method to handle such dual-labeled slices to maintain the integrity of the analysis?
Thank you for your attention to this matter. I appreciate any insights or recommendations you can provide.
Best
Hi @Xiaomin_Lin,
Thank you so much for your post! This is an excellent question, and exactly the kind of behavior that is worth digging deeper into. 
Just to clarify how this dual-labeling occurs in the first place: sct_label_vertebrae involves a straightening step, and the vertebrae are identified in this straightened space (where the cord is parallel to the SI axis, so each slice has a single vertebral label). Then, these straight-space labels are transformed back to the curved space, which naturally causes these “angled” labels. (More details here.)
I think you are right to be concerned about this, especially for the “per level” CSA calculation (sct_process_segmentation -perlevel 1). Since CSA is computed as a slicewise mean within the slices spanned by the vertebral level, I can understand concerns about how such a “partial” slice could potentially affect the mean.
However, tracing the code, it looks like the logic that SCT currently implements for “getting the slices that belong to each vertebral level” is as follows:
In other words, CSA calculations are not done with partial slice areas – the whole slice will be assigned to whatever the “dominant” label is within that slice. (In your screenshot, then, the slice will be included in the slicewise CSA average of the red label, and excluded from the slicewise CSA average of the pink label.)
I hope that clarifies things! 
Kind regards,
Joshua
Thank you for your explanation, Josh. I appreciate the detailed information 
Hi Joshua @joshuacwnewton
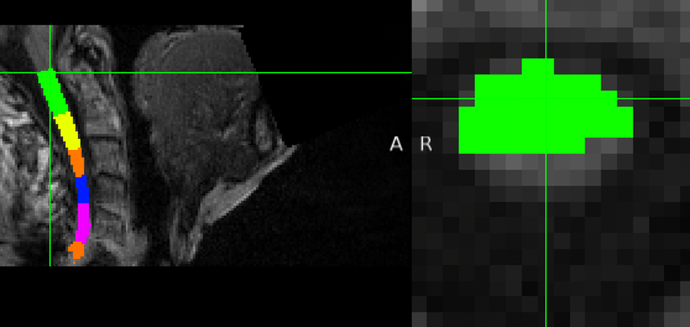
I would like to extend the discussion on the issue I previously raised. Specifically, I have noticed a recurring problem at the edge layers of the output, such as the first layer of C1 and the last layer of C6 (considering my T1 only includes C1~C6 segments). In these layers, the spinal cord voxels are only partially recognized.
Based on your earlier explanation, it seems that these layers, which contain only one value but partial voxels, might still be successfully recognized to some extent. However, upon inspecting the code, I realized that it seems like no mechanism in place to detect this specific scenario.
Given this context, I am concerned about the potential impact this partial voxel recognition might have on the subsequent calculation of CSA for each segment. Could this oversight lead to inaccuracies in the CSA measurements?
Looking forward to your insights on this matter. Thanks!
CSA is computed on the SC seg, not on the vertebral labeled seg. So the file you should be looking at is the SC seg.
Now, about your concern: yes: if a slice shows a partial SC seg, the computed metric will be biased.
How to avoid this? By manually fixing the SC seg at the edge.
BTW: I suggest using the latest, more robust, contrast-agnostic segmentation.