A few comments:
- there is no need to manually crop the image because this step is already included in the
sct_register_to_template function
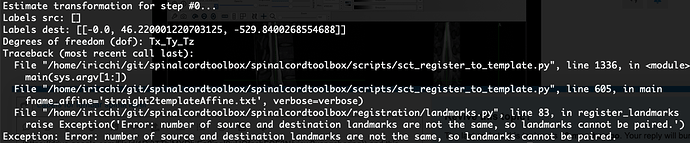
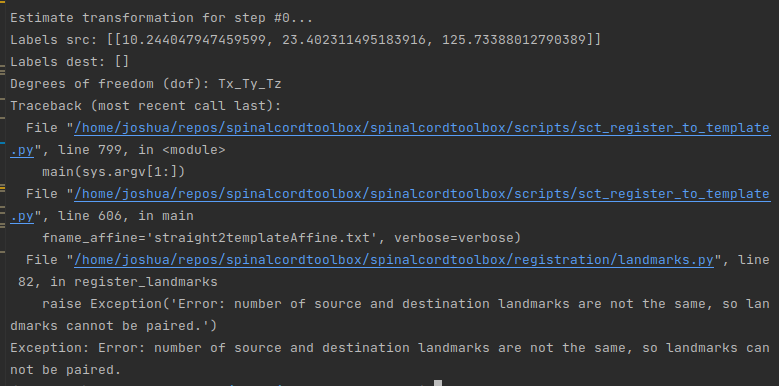
- you cannot use a CSF segmentation with the function
sct_register_to_template unless you specify that the target segmentation used for registration is the CSF mask. This can be achieved with the special flag -s-template-id and selecting the value 6 (you can find this value with the command cat $SCT_DIR/data/PAM50/template/info_label.txt). Although in your case this won’t work because the CSF mask is only the CSF, whereas your CSF mask has the spinal cord and the CSF. So if you want to use this mask you would need to subtract your mask from the SC mask. But in any case, I don’t think this is the right approach for your data. Instead I recommend to stick with the SC segmentation.
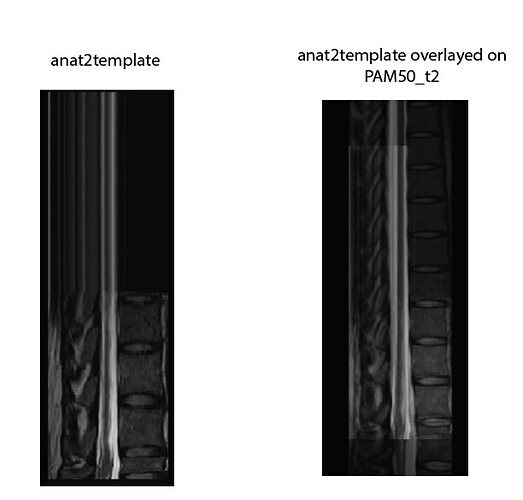
- The artifact you are seeing in Labels for lumbar spinal level - #5 by iricchi is due to the fact that you used more than 2 labels with
sct_register_to_template. When using more than 2 labels, a non-linear registration is first performed (corresponds to the step=0), meaning that SCT will align all the labels, and ignore what’s above the top label and below the bottom label. This results in the strange-looking image that you reported in your post. If you are only interested in the regions between the top and bottom label, then you don’t need to worry about this reconstruction artifact. If however you are interested in the regions outside, then you need to use a maximum of 2 labels.
Here is a piece of code that gets the SC segmentation and only keeps the top and bottom labels from your original label image that you sent (it had 5 labels in total):
# Segment the spinal cord
sct_deepseg_sc -i t2.nii.gz -c t2 -qc qc
# Only keep two labels
sct_label_utils -i labels.nii.gz -keep 17,21 -o labels_17_21.nii.gz
# Register to template
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -ldisc labels_17_21.nii.gz -c t2 -qc qc -param step=1,type=seg,algo=centermassrot:step=2,type=seg,algo=bsplinesyn,metric=MeanSquares,iter=3,slicewise=0:step=3,type=im,algo=syn,metric=CC,iter=3,slicewise=0
It gives the following result, which is acceptable:
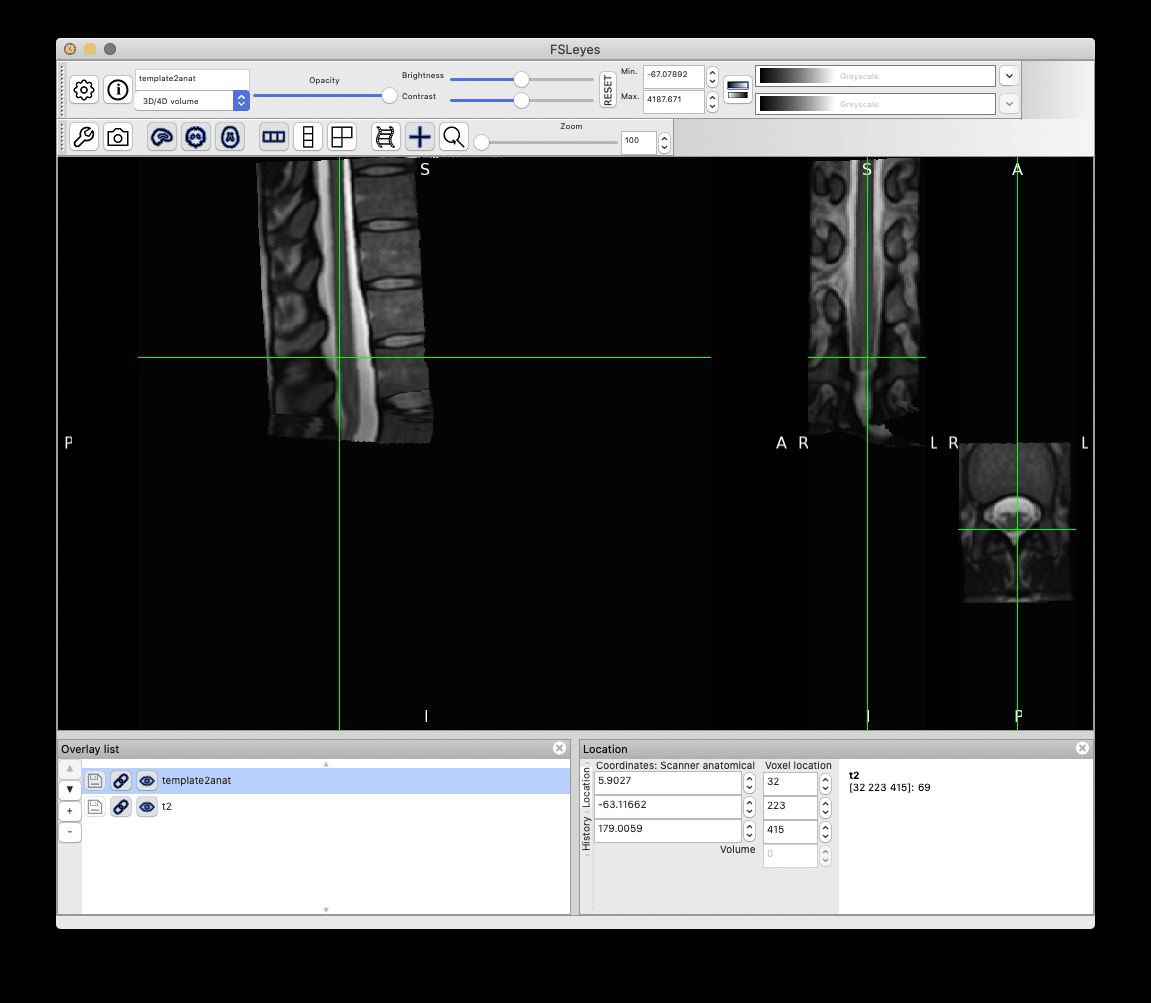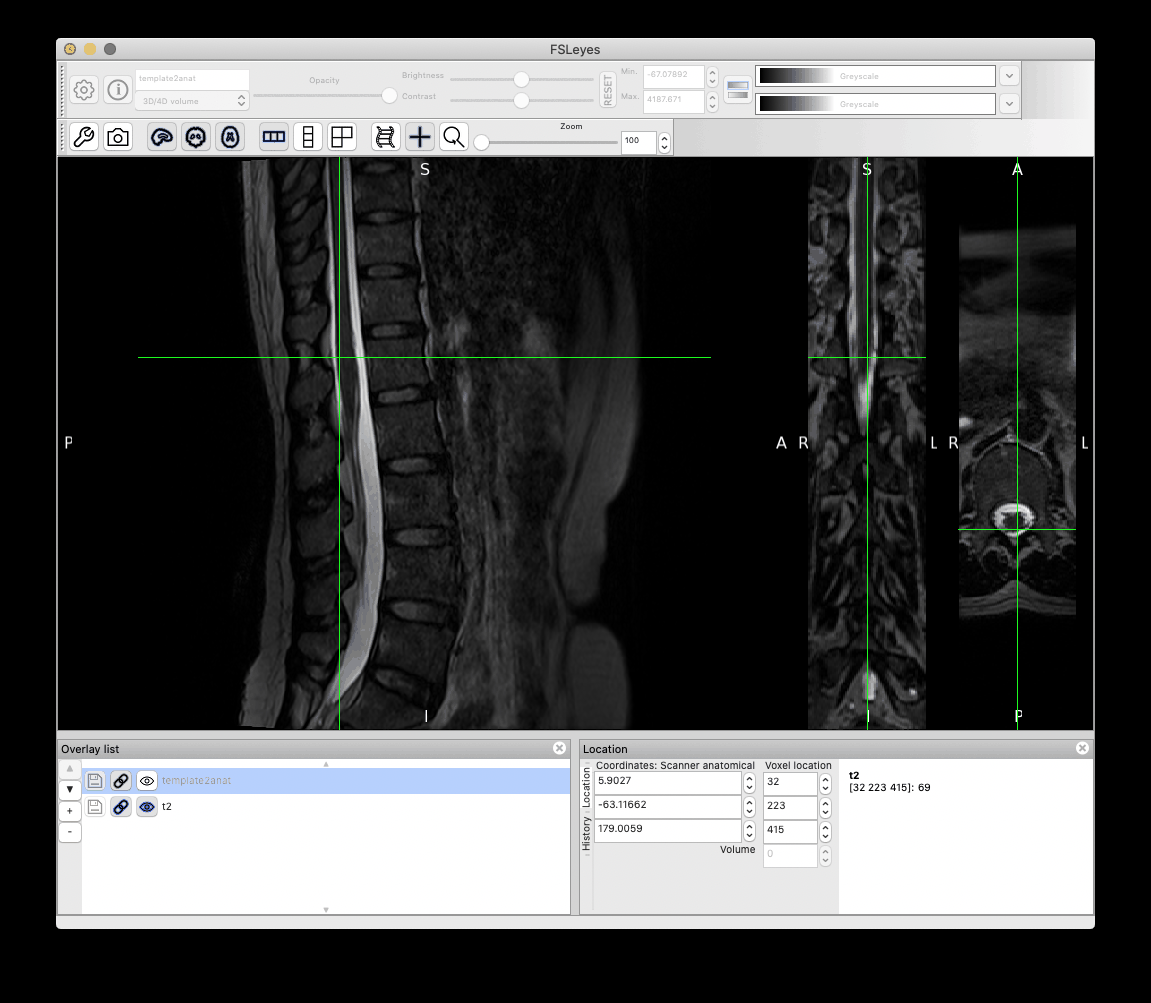
However, as you can notice more clearly in the QC report, the cauda equinea is not perfectly registered with the PAM50 template, because this registration is using intervertebral discs instead of the cord fiducial markers. If you are particularly interested in matching the location of the cauda equinea between your participant data and the PAM50 template, then you can create a label a ‘cauda equinea’ label in the PAM50 template and use that for registration. Demo here:
# Add a label of arbitrary value 99 to the PAM50 template at the start of the cauda equinea
sct_label_utils -i $SCT_DIR/data/PAM50/template/PAM50_label_disc.nii.gz -create-add 70,69,46,99 -o $SCT_DIR/data/PAM50/template/PAM50_label_disc.nii.gz
# Add the same label on the participant data
sct_label_utils -i t2.nii.gz -create 33,239,314,99 -o label_caudaequinea.nii.gz
# Use these labels to register to the PAM50
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -ldisc label_caudaequinea.nii.gz -c t2 -qc qc -param step=1,type=seg,algo=centermassrot:step=2,type=seg,algo=bsplinesyn,metric=MeanSquares,iter=3,slicewise=0:step=3,type=im,algo=syn,metric=CC,iter=3,slicewise=0
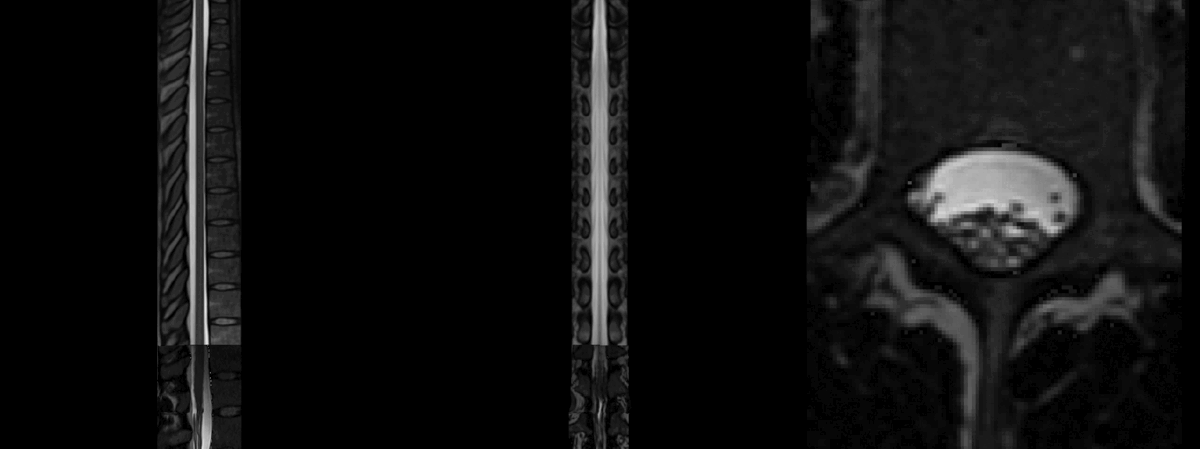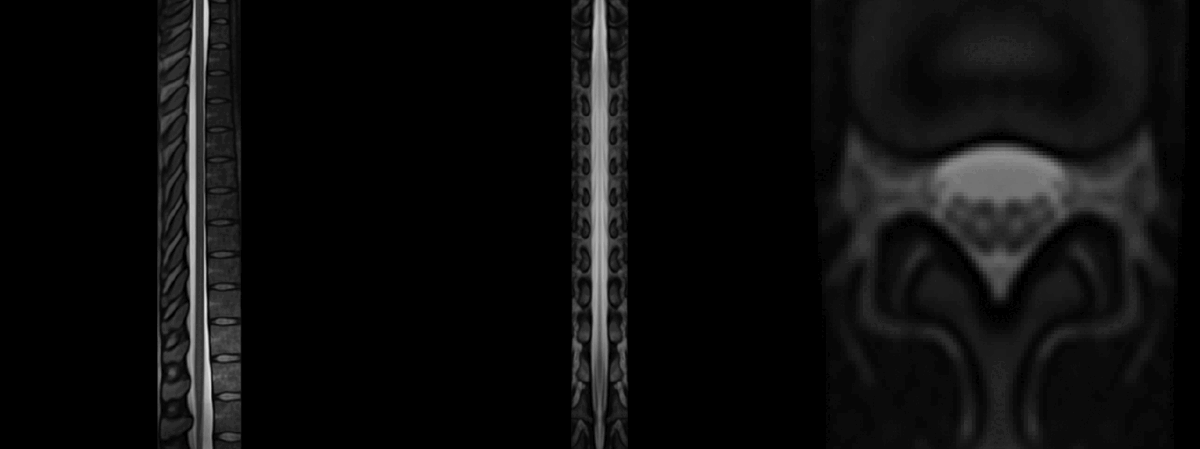
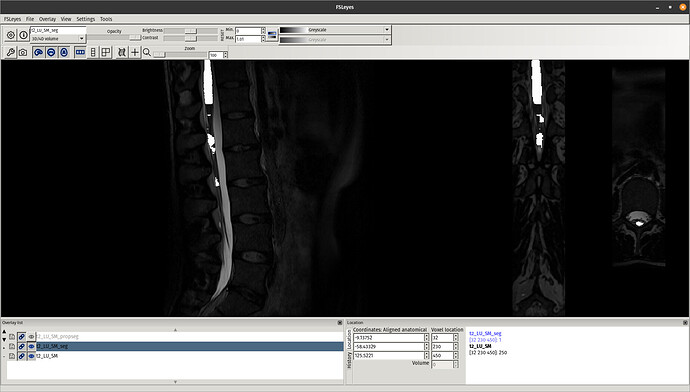
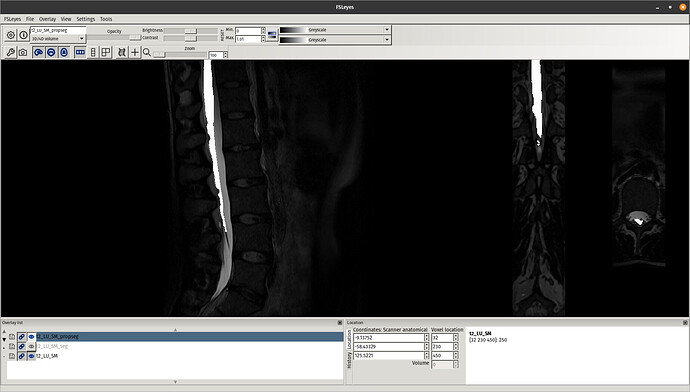
This time, the registration of the lumbar cord is more accurate because now it is based on the cauda equinea (as opposed to the vertebral levels). Notably, the lumbar enlargement is now matching that from the PAM50 better than in the previous registration:
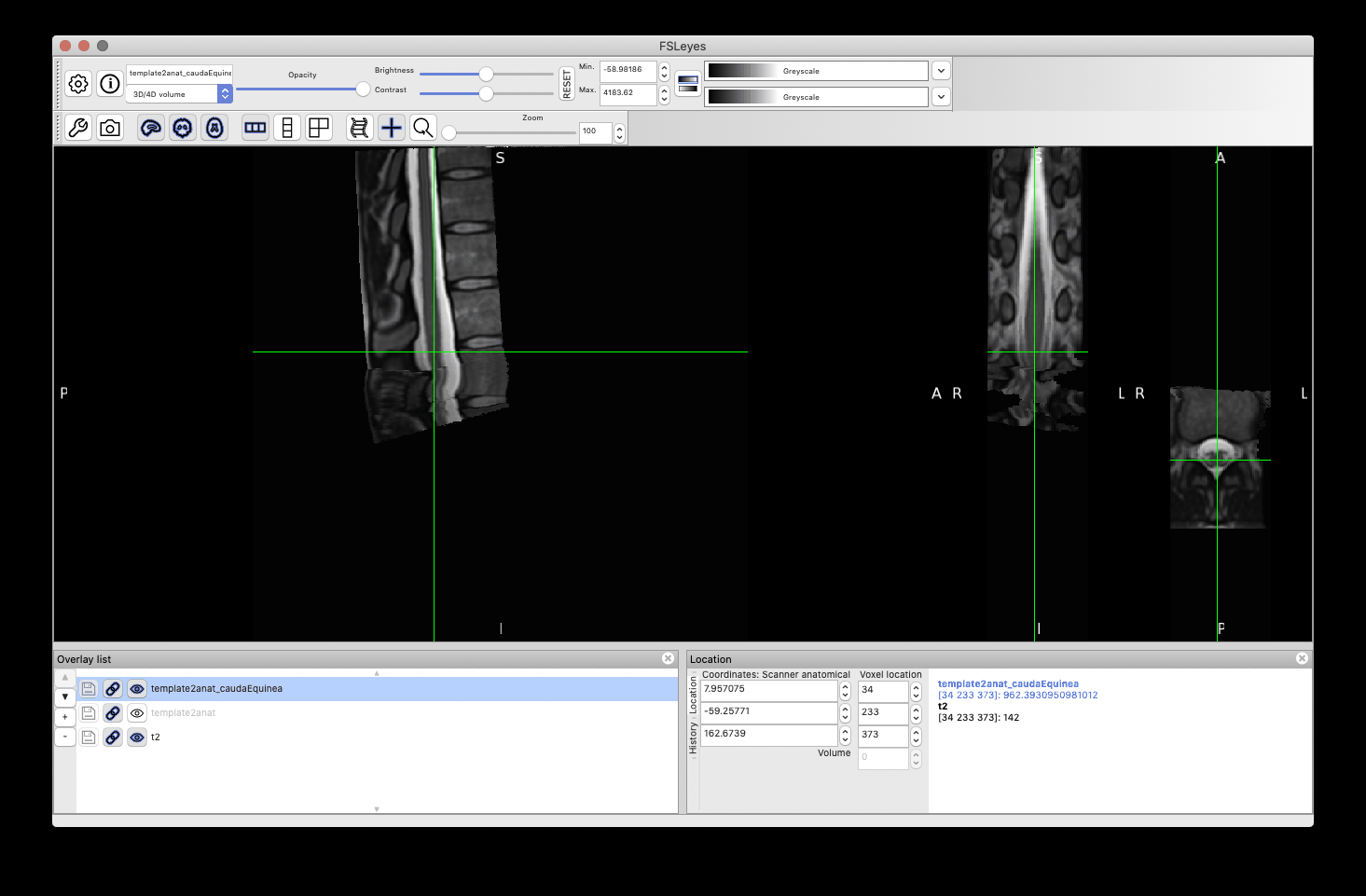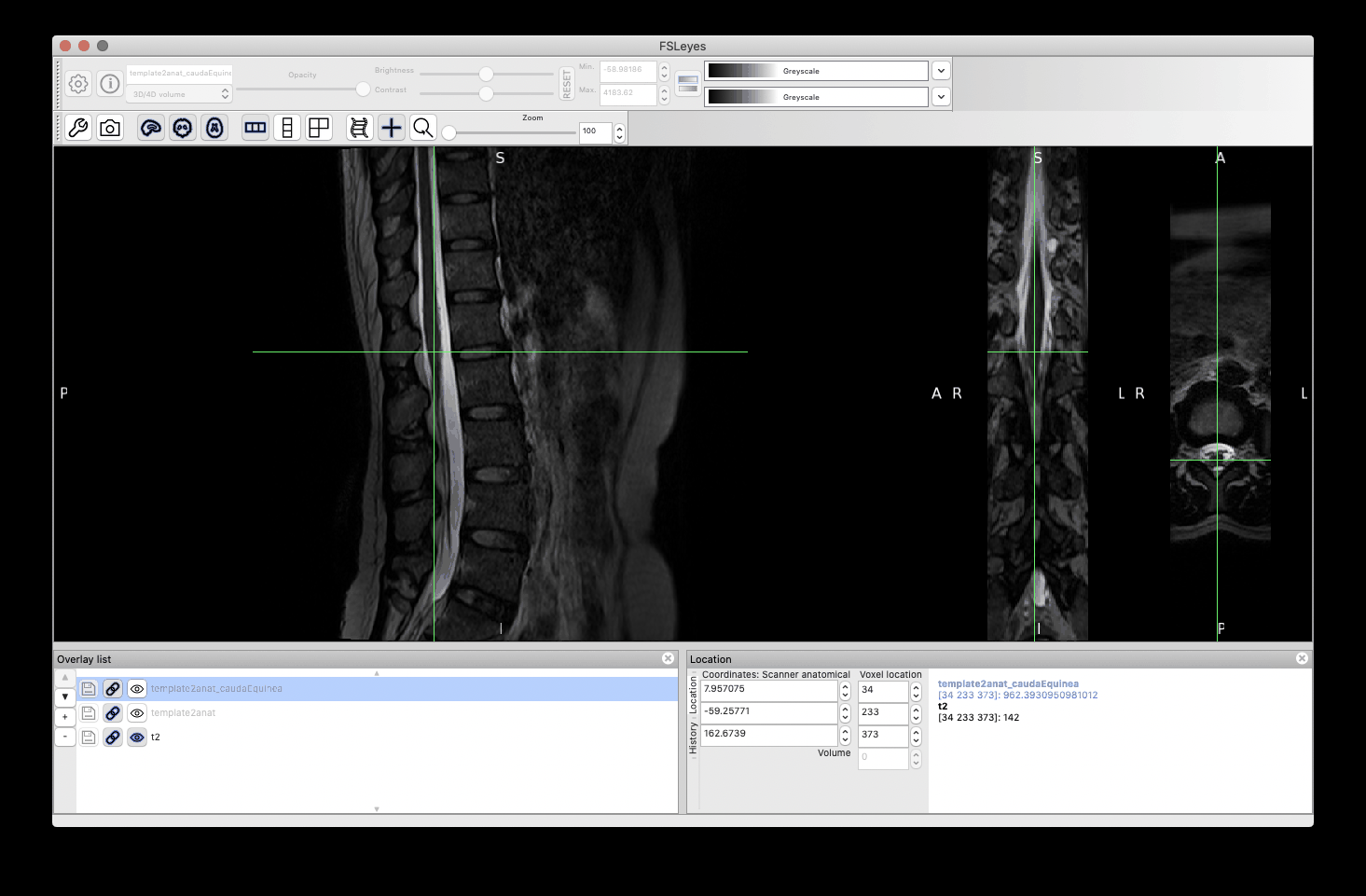
 Notice some image processing artefacts below the cauda equinea, which are caused by the straightening algorithm (because the cord segmentation has “holes” below the cauda equinea). These can be mitigated with a few approaches (eg: extrapolate the cord segmentation, change regularization parameters, etc.), but this is out of the scope of this post. And as mentioned before, if you are only interested in the cord (ie: above the cauda equinea), then that shouldn’t be a problem.
Notice some image processing artefacts below the cauda equinea, which are caused by the straightening algorithm (because the cord segmentation has “holes” below the cauda equinea). These can be mitigated with a few approaches (eg: extrapolate the cord segmentation, change regularization parameters, etc.), but this is out of the scope of this post. And as mentioned before, if you are only interested in the cord (ie: above the cauda equinea), then that shouldn’t be a problem.